

AI Literature Review Generator
Generate high-quality literature reviews fast with ai.
- Academic Research: Create a literature review for your thesis, dissertation, or research paper.
- Professional Research: Conduct a literature review for a project, report, or proposal at work.
- Content Creation: Write a literature review for a blog post, article, or book.
- Personal Research: Conduct a literature review to deepen your understanding of a topic of interest.
New & Trending Tools
Wedding toast generator, ai answer science questions, ai text analyzer and highlights.
How to Use AI for Literature Review (2024): Complete 7 Step Guide for Researchers
Jc Chaithanya
13 min read
What is a Literature Review and Why Does It Matter?
Different types of literature review, method 1: using multiple ai tools for literature review, method 2: using elephas for literature review, advantages of using ai for literature review, the traditional approach: manual literature review, the new kid on the block: ai literature review, comparing the two approaches, common concerns and misconceptions about ai in literature review, things to keep in mind while using ai for literature review, conclusion , 1. can gpt-4 do a literature review , 2. can you use ai to do a literature review, 3. what is the ai tool to summarize the literature review.
Literature reviews are a crucial yet time-consuming part of academic research.
With the advent of artificial intelligence (AI), researchers now have tools that can significantly streamline this process. This guide explores how AI can be effectively utilized to enhance and accelerate literature reviews.
We'll cover the following key aspects:
- The role of AI in literature reviews
- Specific AI tools designed for academic research, such as Elephas
- Best practices for integrating AI into your research workflow
- Potential benefits and limitations of AI-assisted literature reviews
Whether you're a seasoned researcher or a student embarking on your first major project, this guide will provide practical insights on leveraging AI to improve your literature review process.
So let's get started.

When researchers start a new project , they don't just jump in blindly. They first look at what others have already figured out. That's where a literature review comes in handy.
It's like piecing together a puzzle. You gather all the bits of information from different studies, articles, and books. Then you start to see the big picture. What do we know so far? Where are the gaps? Are there any hot debates going on?
All these will translate to
It connects their work to the bigger conversation in their field
It stops researchers from reinventing the wheel
It helps them spot new angles to explore
It shows they've done their homework
By digging into existing research, scholars can push knowledge forward. They're not starting from scratch, but building on what's already there. Plus, a good literature review is super helpful for other researchers too.
It gives them a quick way to catch up on a topic without having to read through piles of separate studies. This saves time and helps guide future research efforts.

Literature reviews can take on various forms depending on their purpose and approach. Below are some of the most popular types of literature reviews:
Narrative Review: This review gives a broad summary of existing studies on a topic but doesn't adhere to a rigid structure. It's often used to provide general insights without analyzing the specifics deeply.
Systematic Review: A systematic review follows a well-defined method to gather, assess, and interpret all available research on a particular question, aiming to reduce bias and provide a more accurate picture of the subject.
Meta-Analysis: This method uses statistical techniques to combine findings from several studies. The goal is to derive a stronger conclusion by merging the data and providing more robust results.
Scoping Review: A scoping review maps out the main ideas and gaps in a field of research, helping to identify where more studies are needed and suggesting potential directions for future research .
Critical Review: This type of review critically examines the strengths and weaknesses of existing research, offering new perspectives or challenging previously accepted theories.
Well, each type offers unique insights based on the research objective, shaping the direction of further inquiry of the research.
Step-by-Step Guide on How to Use AI for Literature Review
Using AI for literature review can significantly streamline your research process. Let's explore two methods: a multi-tool approach and using Elephas, an all-in-one assistant.

1. Identify Your Research Topic and Keywords
The first step is defining your research area. Use Perplexity AI for topic exploration and generating research questions. This AI tool helps you uncover new angles for research topics by analyzing vast amounts of data quickly.
2. Search for Relevant Articles
Start your literature search by heading to Elicit.org. Export the articles based on categories like abstract, author, title, and publication date. You can also use other AI-powered search tools like Semantic Scholar or Google Scholar to broaden your sources.
3. Generate Summaries and Key Themes with GPT-4o
After gathering your articles, use GPT-4o to analyze the abstracts and generate key themes. Input the abstracts with prompts like, “ Please summarize the key themes from these articles. ” This step saves hours of manual reading and gives you a thematic overview.
4. Draft an Initial Literature Review with Copy.ai
Use Copy.ai to create a first draft of your literature review. Its AI-powered writing features allow you to generate sections of the review quickly and in a structured format. Copy.ai can assist in writing specific sections, such as background or methodology, based on the keywords and themes you provide.
5. Refine with Smart Writing Tools
Use AI tools like Jasper or Writesonic to refine your literature review. These tools help to paraphrase content, improve readability, and adapt the tone to meet academic standards. The rewrites from these tools can help make the content more engaging and coherent.
6. Organise References Using Reference Managers
As you finalise your draft, integrate reference management tools like Mendeley or Zotero. These tools can store and organise all the references cited in your paper, and AI integration allows for easy reference generation.
7. Use Perplexity AI for Final Checks
Before submission, use Perplexity AI again to check for any gaps in the research or identify potential new areas to explore. It can provide suggestions based on the latest publications and research trends.

Elephas offers an all-in-one solution for conducting literature reviews. Here’s how you can use it:
1. Search the Web with Elephas
Elephas’ web search feature allows you to find relevant articles directly within the tool. Simply input your research terms, and it will pull up related papers, articles, and sources for you to analyse.
2. Analyse Key Themes
Using integrated AI models like GPT-4 or Claude, you can analyse the abstracts or summaries of these papers to identify key themes.
3. Generate a Literature Review
With Elephas’ Smart Write feature, you can create a well-structured literature review in just a few prompts. It pulls in the key themes and drafts a coherent review, ensuring that all relevant abstracts are referenced accurately.
4. Organise with Super Brain
Elephas’ Super Brain feature helps you manage the knowledge from the papers, documents, and research you’re using. It organises and categorises the data for easy access during the writing process.
5. Refine and Customise Tone
Elephas allows you to refine the review by using its multiple writing modes (Zinsser, Friendly, Professional, or Viral Mode). You can ensure that the literature review matches your preferred tone and style.
6. Manage References and Citations
With the help of Super Brain, you can manage references and citations within the text, simplifying the process of creating a bibliography.
By using Elephas, you can significantly speed up the literature review process while maintaining high quality. It’s a comprehensive all-in-one AI tool, making it one of the best solutions for conducting literature reviews.

AI is changing the game for researchers tackling literature reviews. Now, we've got smart tools that can do a lot of the heavy lifting for us. Let's explore some advantages of using AI for literature review.
Time Efficiency: AI dramatically speeds up the review process. It can analyze thousands of articles in minutes, a task that would take humans days or weeks to complete.
Comprehensive Coverage: AI can thoroughly scan vast databases, ensuring no relevant study slips through the cracks.
Pattern Recognition: AI literature tools excel at identifying trends and connections across multiple studies, often spotting insights that humans might overlook.
Bias Reduction: AI approaches each piece of literature objectively, helping to minimize human biases that can creep into manual reviews.
Multilingual Capabilities: Language barriers become less of an issue. AI can process and analyze research in multiple languages, broadening the scope of reviews.
Data Visualization: Many AI tools can generate clear, insightful visualizations of complex data, making it easier to grasp key findings at a glance.
Continuous Updating: In rapidly evolving fields, AI can keep literature reviews current by continuously incorporating newly published research.
While AI brings these impressive benefits to the table, it's important to remember that it's not a wise move to use AI extensively and limit your human touch in the review.
The ideal approach is to combine AI with your critical thinking and domain knowledge. As AI technology continues to advance, its role in streamlining and improving literature reviews is only set to grow, opening up exciting new possibilities for more comprehensive and efficient research processes.
Manual Literature Review vs AI Literature Review
In the world of research, literature reviews play a crucial role. They help researchers understand what's already known about a topic and identify gaps in knowledge. Today, we're seeing a shift in how these reviews are conducted, with AI tools coming in and helping researchers to reduce their overall workflow. But what is actually better: manual literature review or AI-assisted literature reviews?

Manual literature reviews have been the standard for a long time. Here's what they typically involve:
They take detailed notes on each source
Researchers spend hours reading through papers and articles
Key themes and patterns are identified through careful analysis
Connections between different studies are made based on the expertise
This method has its strengths. It allows for deep understanding and critical thinking. Researchers can pick up on subtle nuances that might be important. However, it's also very time-consuming and can be limited by the researcher's ability to process large amounts of information.

AI-assisted literature reviews are changing the game. Here's how they work:
AI tools can quickly scan thousands of articles
Key themes and patterns are automatically extracted
They use advanced algorithms to identify relevant studies
Connections between studies are made based on data analysis
The speed and efficiency of AI reviews are impressive. They can process far more information than a human could in the same amount of time. This means researchers can get a broader view of their field quickly. AI tools are also great at spotting trends and connections that humans might miss.
When we look at manual and AI literature reviews side by side, we see some interesting differences:
Time efficiency: AI is much faster, potentially saving weeks of work
Scope: AI can cover a broader range of sources
Depth of analysis: Manual reviews often provide deeper insights
Bias: AI can help reduce human bias, but may have its own algorithmic biases
Flexibility: Manual reviews can adapt more easily to unique research needs
Language: AI can work across multiple languages, expanding the scope of research
It's important to note that AI isn't perfect. It might miss context or nuances that a human would catch. That's why many researchers are now using a hybrid approach.
They use AI to do the initial heavy lifting, then apply their own expertise to refine and interpret the results.
In the end, whether manual or AI-assisted, the goal of a literature review remains the same: to build a solid foundation for new research and contribute to the advancement of knowledge in the field.

There are many concerns and misconceptions about using AI in literature reviews. It's natural to have doubts about new technology, especially when it comes to something as crucial as research.
One big worry is that AI might replace human researchers. But that's not really the case. AI is a powerful tool, but it can't match the critical thinking and deep understanding that humans bring to the table, at least for now. It's more of a helper than a replacement.
Accuracy Concerns: There's a misconception that AI might misinterpret or miss important information. Modern AI tools are actually quite accurate, but they do need proper setup and oversight to perform at their best.
Over-reliance on Technology: Some worry researchers might become too dependent on AI, losing their own analytical skills. In reality, AI frees up time for deeper analysis and creative thinking.
Data Privacy Issues: Concerns about data security and privacy are valid. It's crucial to use AI tools that adhere to strict data protection standards.
Limited to Quantitative Analysis: Many think AI can only handle numbers and statistics. Actually, advanced AI can process qualitative data too, including complex text analysis.
High Costs: While some AI tools can be expensive, many affordable options exist. The efficiency gains often outweigh the initial investment.
Complexity of Use: There's a belief that AI tools are too complicated for the average researcher. In fact, many are designed with user-friendly interfaces.
When using AI for literature reviews, there are several key points to keep in mind. Let's check out these important considerations that might get you into trouble if not checked properly when using AI for a literature review.
Quality Control: While using AI, you need to always double-check the results it generates. Take the time to review the selected articles and ensure they're truly relevant to your research.
Ethical Considerations: The use of AI in academic work is still a hot topic. Be mindful of ethical concerns, particularly around plagiarism and AI-generated content. Make sure your work is original and properly cited.
Stay Updated: Keep an eye on the latest developments in AI tools for literature reviews. What's new in the AI market and what’s outdated will help inform you to make the most of these tools.
Define Clear Parameters: Be specific about your research questions, keywords, and inclusion criteria. The more precise your input, the more relevant your results will be.
Understand AI Limitations: AI is great at processing large amounts of data, but it might miss nuances or context that a human would catch.
Maintain a Critical Perspective: Don't accept AI-generated summaries or analyses at face value. Apply your critical thinking skills. Question the results, look for potential biases, and consider alternative interpretations.
Document Your Process: Keep detailed records of how you used AI in your review. Note which tools you used, what parameters you set, and how you verified the results. This transparency is vital for the credibility of your work.
These tips may be generic and known to everyone, but many researchers, while using AI in their literature writing or revising process, still make these mistakes. Using AI is not wrong, but it's about finding the right balance between technological assistance and human expertise.
You know, it's pretty amazing how AI is shaking things up in the world of research. If you're knee-deep in literature reviews, learning to use AI could be a game-changer for you. It's like having a super-smart assistant who never gets tired and can spot connections you might miss.
There are a bunch of ways to go about it - you could mix and match different AI tools, or go for an all-in-one solution. The trick is finding what clicks for you. Just remember, AI is incredibly helpful, but it's not all good. You've still got to bring your expertise to the table.
As AI keeps evolving, it's opening up new possibilities for research. Who knows what breakthroughs we might see? So, getting comfortable with AI for literature reviews now could really set you up for the future.
It's an exciting time to be a researcher, that's for sure!
Yes, GPT-4 can help with a literature review by summarizing research papers, analyzing content, and identifying key themes. It speeds up the process by offering relevant insights from sources, but human expertise is still needed to ensure accuracy and a comprehensive understanding.
Yes, AI can assist in conducting a literature review by automating tasks such as summarizing research papers, analyzing large amounts of data, and highlighting important findings. This aids in streamlining the review process, although human judgment is essential for interpreting and validating the results effectively.
AI tools like Elephas, designed for summarizing literature reviews, help streamline the process by providing features such as offline support, multiple language models, and web search integration. These tools can quickly summarize key insights and trends across academic papers and other research sources.
AI assistant
Sign up now
Get a deep dive into the most important AI story of the week. Deliverd to your inbox for free!

Meet Elephas - Your AI-Powered Knowledge Assistant. Your Personal ChatGPT for all your files. Transform information overload into actionable insights. Organize vast knowledge. Access ideas efortlessly. Save 10 hours a week
You may also want to read

The Growing Impact of AI on US Elections: Opportunities, Threats, and Safeguards
Pinned Post

Anthropic Introduces Claude 3.5 Sonnet Update and Revolutionary Computer Use Capability

Top 10 Best ChatGPT (AI) Apps for Essay Writing (2024) | Free + Paid
Previous Post

AI Tools To Automate Your Literature Review: Which To Use?
Researching and writing outa literature review can be a daunting task, but AI-powered tools like Semantic Scholar, Research Rabbit, and Scite are revolutionising this process.
These tools, leveraging artificial intelligence, automate the arduous task of sifting through mountains of academic papers, extracting key information, and summarizing relevant research.
In this article, lets explore how AI research assistants and advanced search engines streamline the literature review process, making it faster, more efficient, and thorough for conducting scientific research. Discover the best AI tools to transform your literature review work.
Why Use AI Tools To Write Literature Review?
The vast sea of academic papers can be daunting. This is where AI tools like ChatGPT, Semantic Scholar, and Research Rabbit become indispensable.

AI tools for literature review are adept at sifting through millions of papers across various databases like PubMed and Google Scholar. They use AI algorithms and natural language processing (NLP) to identify relevant research articles, providing a summary of key information.
Some AI-powered research assistant such as Scite, uses AI to scan through research data, offering insights into the supporting or contrasting evidence within peer-reviewed papers. This can be a valuable tool for finding specific details in a systematic literature review.
These tools also streamline the literature search by automating the process of citation and reference management. Tools like Connected Papers and Research Rabbit are adept at evaluating and summarizing relevant academic papers, saving you time and effort.
They can extract key information from PDF documents and research articles, making it fast and easy to organize your findings.
AI-powered tools for literature review also help in creating a thorough literature search, essential for:
- Systematic literature reviews
- Meta-analyses, and
- Identifying research gap.
They can elicit information from a variety of sources, providing a comprehensive view of your research question. This approach ensures that your literature review is not only rich in content but also diverse in perspectives.
Best AI Tools To Write Literature Review
If you are looking at exploring AI tools to help you write literature review, consider these instead. They are revolutionizing the way literature reviews are conducted:
Connected Papers
As the name suggests, Connected Papers focuses on showing how various academic papers are interconnected. This generative AI tool helps you visualize the research landscape around your topic, making it easier to:
- Identify key papers,
- Identify potential gaps in your literature review, and
- Understand emerging trends in scientific research.
While not a traditional literature review tool, ChatGPT is revolutionising academic research by offering prompt-based assistance.
This chatbot-like AI tool can help automate parts of the literature review process, such as generating research questions or providing quick summaries of complex topics.
ChatGPT’s AI-driven insights can be a valuable starting point for deeper exploration into specific research areas. The key is to provide it with the right input, and then to give the right prompts.
Research Rabbit
This tool uses AI to scan a variety of sources, including peer-reviewed research data. It’s particularly useful for conducting systematic literature reviews, as it can evaluate and summarize relevant academic papers, highlighting supporting or contrasting evidence.

Its key features include mapping out connections between research papers, identifying key papers in a research area, and helping to understand how various research topics are interconnected.
Research Rabbit also excels in reference management, a vital component of academic writing.
Scite is an innovative tool that uses AI technologies to provide a new layer of insight into scientific literature.
It evaluates the credibility of research by analyzing citation contexts, helping you to identify the most impactful and relevant papers for your research question.
Scite’s most distinctive feature is its Smart Citations. Unlike traditional citations that merely count how often a paper is cited, Smart Citations provide context by showing how a paper has been cited. This means that for each citation, Scite shows you the other relevant papers that:
- Contrasts, or
- Mentioning evidence for the claims made in the original paper.
As a result, Scite is invaluable for conducting thorough literature reviews, especially for systematic reviews and meta-analyses.
Semantic Scholar
This AI-powered research assistant stands out in its ability to sift through vast databases like PubMed and Google Scholar.
It uses advanced natural language processing (NLP) and AI algorithms to extract key information from millions of academic papers, offering concise summaries and identifying relevant research articles.
What sets Semantic Scholar apart is its AI algorithms. It analyses millions of academic publications, extracting key information such as:
- Figures,
- Tables, as well as
- Contextual relevance of each paper.
This enables the tool to provide highly relevant search results, summaries, and insights that are tailored to your specific interests and research needs.
What To Watch Out For When Writing Literature Review With AI Tools?
When you’re writing a literature review with AI tools, you’re stepping into a world where technology meets academic rigor. To ensure they balance each other out, by watching over these details as you use AI when researching:
Bias
AI algorithms, including those used in literature search tools, can inherit biases from their training data.
This might skew the search results towards more popular or frequently cited papers, potentially overlooking lesser-known yet significant research.
Ensure you’re accessing a variety of sources to maintain a balanced perspective. When possible, always look into newer papers on your research area, as these may not have been discovered by your AI tool yet.
Citation Accuracy
While tools like Scite provide advanced citation analysis, the accuracy of AI in identifying and interpreting citations is not infallible.

Always verify the citations and references manually, especially when dealing with complex literature or less digitised sources.
You can also use multiple AI tools to check for citation accuracy. For example, rather than simply relying on Scite, you can also use other reference management softwares like:
- Refworks, or
Contextual Understanding
AI tools, efficient as they are, might not fully grasp the nuanced context of your research question.
Tools like ChatGPT and Semantic Scholar provide summaries and identify relevant papers using natural language processing (NLP), but they may not always align perfectly with your specific research focus.
Always double-check that the AI’s interpretation matches your intended research angle. This means actually spending time to read articles within the research area, and become familiar with it.
Depth of Analysis
AI tools can automate the initial stages of your literature review by quickly sifting through databases like PubMed and Google Scholar to find relevant papers.
However, they might not evaluate the depth and subtlety of arguments in academic writing as thoroughly as a human researcher would.
It’s crucial to supplement AI findings with your own detailed analysis. This means you should not rely on AI completely, but to actually roll up your sleeves and work on the analysis yourself as well.
Over-reliance on AI
There’s a temptation to overly rely on AI for streamlining every aspect of the literature review process.

Remember, AI is a tool to assist, not replace, your critical thinking and scholarly diligence.
Use AI to enhance your workflow but maintain an active role in evaluating and synthesising research data. See AI as a tool, not your replacement. You need to be there to pilot the AI tool, to ensure it is doing its job properly.
AI Tools For Literature Review: Keep Up With Times
Leveraging AI tools for your literature review is a game-changer. Tools like Semantic Scholar, Research Rabbit, and Scite, powered by advanced AI algorithms, not only automate the search for relevant papers but also provide critical summaries and evaluations.
They enhance the literature review process in academic research, making it more efficient and comprehensive.
As AI continues to evolve, these tools become indispensable for researchers, helping to streamline workflows, organize findings, and extract key insights from a vast array of scientific literature with unparalleled ease and speed.

Dr Andrew Stapleton has a Masters and PhD in Chemistry from the UK and Australia. He has many years of research experience and has worked as a Postdoctoral Fellow and Associate at a number of Universities. Although having secured funding for his own research, he left academia to help others with his YouTube channel all about the inner workings of academia and how to make it work for you.
Thank you for visiting Academia Insider.
We are here to help you navigate Academia as painlessly as possible. We are supported by our readers and by visiting you are helping us earn a small amount through ads and affiliate revenue - Thank you!

2024 © Academia Insider

A free, AI-powered research tool for scientific literature
- Thomas Devereaux
- Knot Theory
New & Improved API for Developers
Introducing semantic reader in beta.
Stay Connected With Semantic Scholar Sign Up What Is Semantic Scholar? Semantic Scholar is a free, AI-powered research tool for scientific literature, based at Ai2.

The Guide to Literature Reviews

- What is a Literature Review?
- The Purpose of Literature Reviews
- Guidelines for Writing a Literature Review
- How to Organize a Literature Review?
- Software for Literature Reviews
- Introduction
Why use artificial intelligence for a literature review?
How can ai assist in literature reviews.
- Which AI-powered tools can be used for literature reviews?
Future research directions of AI-powered tools for literature reviews
- How to Conduct a Literature Review?
- Common Mistakes and Pitfalls in a Literature Review
- Methods for Literature Reviews
- What is a Systematic Literature Review?
- What is a Narrative Literature Review?
- What is a Descriptive Literature Review?
- What is a Scoping Literature Review?
- What is a Realist Literature Review?
- What is a Critical Literature Review?
- Meta Analysis vs. Literature Review
- What is an Umbrella Literature Review?
- Differences Between Annotated Bibliographies and Literature Reviews
- Literature Review vs. Theoretical Framework
- How to Write a Literature Review?
- How to Structure a Literature Review?
- How to Make a Cover Page for a Literature Review?
- How to Write an Abstract for a Literature Review?
- How to Write a Literature Review Introduction?
- How to Write the Body of a Literature Review?
- How to Write a Literature Review Conclusion?
- How to Make a Literature Review Bibliography?
- How to Format a Literature Review?
- How Long Should a Literature Review Be?
- Examples of Literature Reviews
- How to Present a Literature Review?
- How to Publish a Literature Review?
Using Artificial Intelligence for Literature Reviews
Literature reviews are known to be time-consuming. From finding the articles to reading and analyzing all the components of the publications, it can take a long time to conduct a literature review. However, AI-based tools can help researchers speed up the process.
In recent years, using AI for literature reviews has gained significant importance in academic research. Specifically, AI tools have revolutionized how scholars and students explore various sources, offering cutting-edge capabilities to fine-tune search terms and sift through full-text articles with ease. For instance, AI-powered suggestions help researchers identify relevant scholarly articles and research papers, streamlining the development of a thorough literature review. Tools like Semantic Scholar and Connected Papers provide insightful citation contexts and connect related research which saves time and adds detail and precision in summarizing the breadth of existing knowledge.
Conducting a literature review involves summarizing and discussing published ideas from various sources to build a comprehensive view of the current state of research. AI can significantly enhance this process by offering meta-analysis capabilities and helping researchers write summaries that encapsulate the essence of multiple publications. For example, AI can analyze PDF documents and extract key points that help you to write more concise and informative summaries. While all researchers should take responsibility for any AI-generated outputs they end up using in their research you can efficiently navigate through the sea of academic work using AI and save time.
AI's ability to assist in literature reviews extends beyond just finding and summarizing articles. It can also play a pivotal role in the early stages of research projects by helping to refine research questions and identify gaps in the existing literature. For example, AI tools can scan a wide array of scholarly articles to identify emerging trends and suggest areas that warrant further exploration.
According to recent research, AI can help with problem formulation and identify and verify research gaps by analyzing large datasets (Wagner, Lukyanenko, & Paré, 2021). However, human judgment is crucial for nuanced and context-specific interpretations. While AI can significantly reduce the time and effort required for repetitive tasks in literature reviews, human interpretation and creativity are irreplaceable for insightful synthesis and theory development.
These tools have demonstrated considerable potential in automating and streamlining the literature review process, especially in the critical phases of screening and data extraction. However, several limitations need to be addressed, particularly regarding usability and the integration of more advanced AI technologies (Bolaños, Salatino, Osborne, & Motta, 2024). The usability issues, such as a steep learning curve and misalignment with user requirements, hinder the widespread adoption of these tools among researchers. Financial constraints also pose a barrier, limiting access to comprehensive functionalities in some cases.
Nonetheless, by integrating these AI-powered tools into the literature review process, researchers can enhance their efficiency, reduce the cognitive load associated with manual tasks, and focus more on the interpretative and theoretical development aspects of their work. The continuous development and improvement of these tools promise even greater support for researchers in the future, making literature reviews more manageable and insightful (Wagner, Lukyanenko, & Paré, 2021).

Finding literature
AI-powered tools like machine learning algorithms and natural language processing (NLP) can significantly streamline finding relevant literature. These tools can search extensive databases and digital libraries to identify articles, books, and papers that match specific keywords and topics. They can also recommend related works based on the content of already identified materials, saving researchers considerable time and effort.
Literature summaries
AI can assist in summarizing large volumes of literature quickly and accurately. Tools using natural language processing (NLP) can extract key points, themes, and findings from numerous articles and create concise summaries. This helps researchers grasp the essential information from a broad array of sources without having to read each one in full detail. Such summarization tools can be invaluable for literature reviews, providing a clear overview of the existing research landscape.
Analyzing text
AI can perform in-depth text analysis, identifying patterns, trends, and insights that might not be immediately apparent. Techniques such as sentiment analysis , topic modelling, and keyword extraction can reveal underlying themes and connections within the literature. This analytical capability allows researchers to draw more nuanced conclusions and identify gaps or areas for further study.

Screening for inclusion
During the literature review process, AI can assist in screening articles for inclusion based on predefined research criteria. Machine learning models can be trained to recognize relevant studies by analyzing abstracts and full texts, thereby automating the initial screening phase. This reduces the manual workload and ensures a more systematic selection process.
Quality assessment
AI's potential in quality assessment is moderate, as this step requires expert judgment to evaluate methodological rigour. Tools equipped with AI can provide initial insights about the quality of other studies by checking for common issues related to how transparently methods are described and verification checks authors conducted. By automating parts of the quality assessment, researchers can focus on more in-depth analysis and interpretation of high-quality studies.
Information extraction
AI tools can streamline the data extraction process by automatically identifying and extracting relevant data points from research articles. Machine learning algorithms can be trained to recognize and pull out specific types of information, such as statistical results, sample characteristics, and key findings. Tools equipped with AI can provide initial insights about the quality of other studies by checking for common issues related to how transparently methods are described and verification checks authors conducted.
Ready to try out ATLAS.ti?
Intuitive AI tools to help you with your research projects. Check them out with a free trial of ATLAS.ti.
AI tools are designed to handle large volumes of data efficiently, making the literature review process faster and more comprehensive. With the emergence of the tools, researchers are exposed to several tools they can use. From summarizing articles to assisting in the literature search, AI tools can assist researchers by saving time and providing new ways of analyzing data.
For example, Research Rabbit maps research papers so that researchers can visualize connections between papers, authors and topics. Some research discovery tools such as LitSonar facilitate database searches by automatically translating search queries into formats compatible with various literature databases, such as EBSCO , AIS eLibrary , and ProQuest .
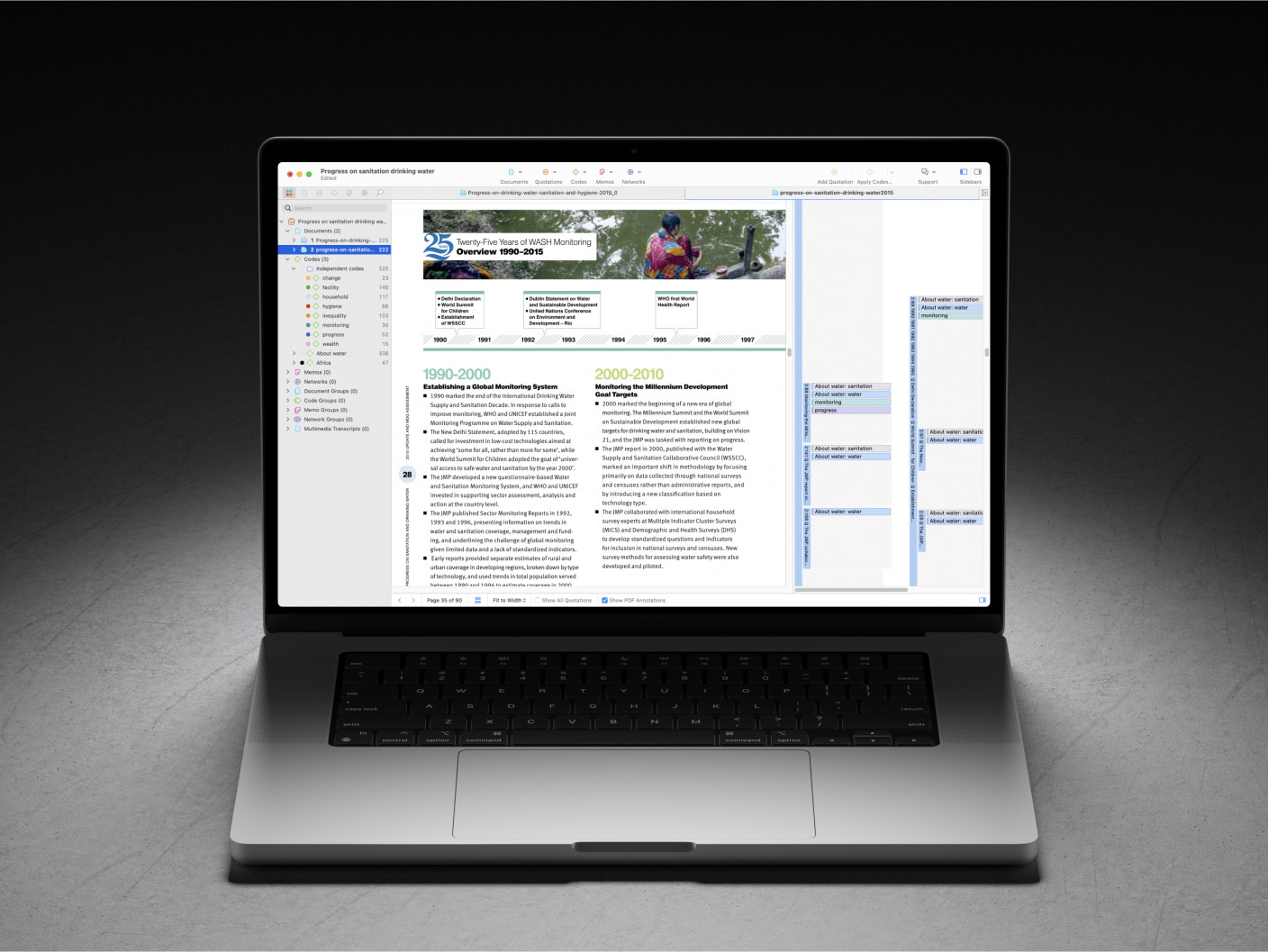
ATLAS.ti is a comprehensive CAQDAS that has incorporated a suite of AI tools that can facilitate literature reviews. This includes different AI functions to find literature, summarize, and code it.
Paper Search in ATLAS.ti Web
Paper Search 2.0 in ATLAS.ti Web is a cutting-edge AI tool that streamlines research workflows by quickly assessing the relevance of scientific papers. Users input their research questions, and the tool performs comprehensive searches, delivering concise summaries of the most pertinent papers. This innovation saves researchers time and effort, allowing them to focus on analysis.
Paper Search 2.0 accesses over 200 million scientific resources from Semantic Scholar , providing an extensive database for relevant studies. Its streamlined search function efficiently finds and imports key scientific resources. The AI offers focused summaries of the top five papers, highlighting content relevant to the user’s research question. Additionally, it allows for easy citation of documents within ATLAS.ti projects, ensuring a fully integrated workflow.
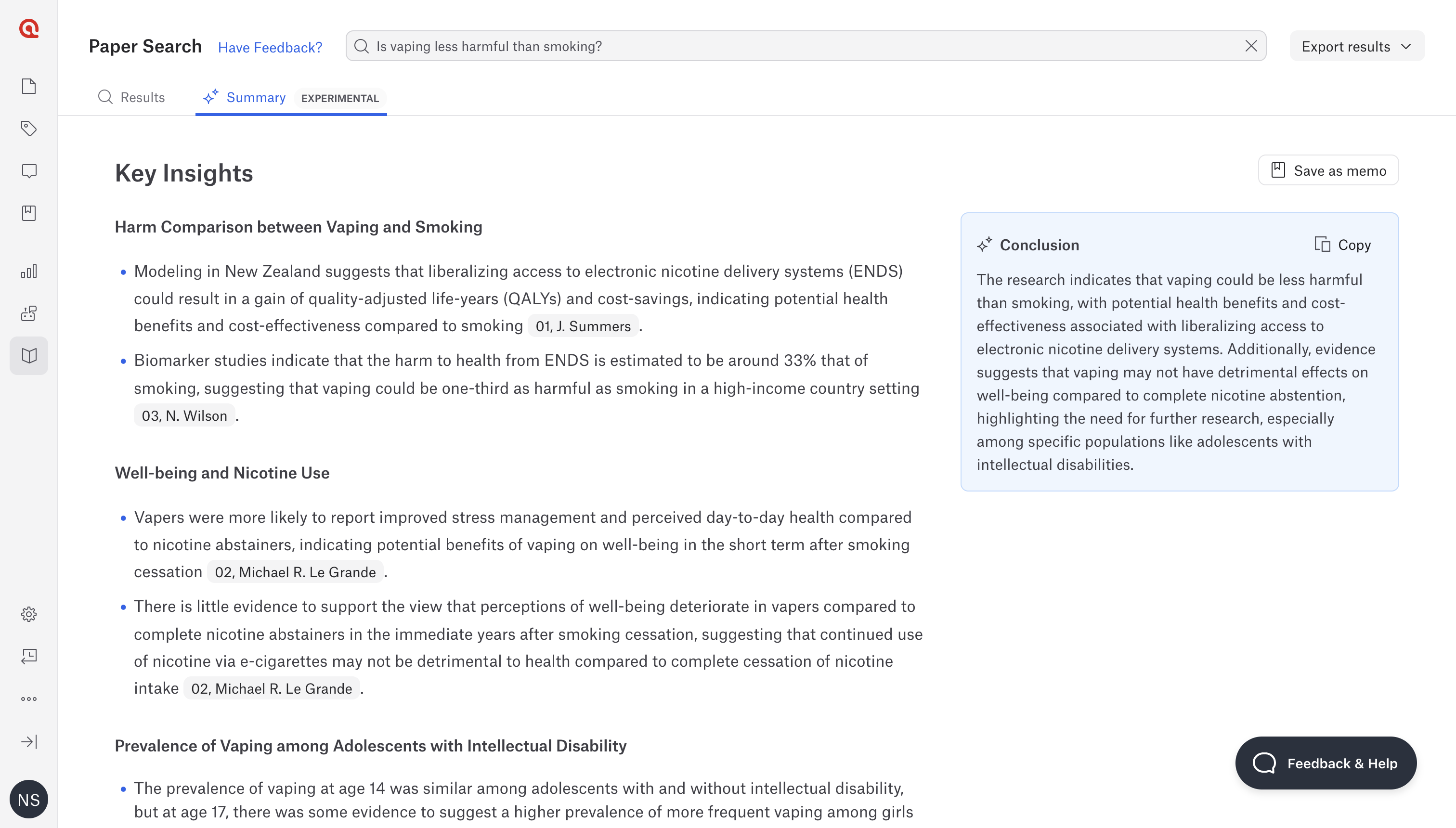
The tool supports the entire literature review process within a single platform. Advanced Natural Language Processing ensures a deep understanding of research needs, resulting in highly relevant search results and summaries. When users input their research questions, the AI refines them, generates relevant keywords, and searches Semantic Scholar’s database. It identifies pertinent papers, summarizes findings, and delivers actionable insights. This process, driven by the ATLAS.ti AI Lab, transforms raw data into valuable knowledge, ensuring researchers receive key insights applicable to their work.
ATLAS.ti Web enables researchers to complete their literature review using AI tools and qualitative data analysis features. Targeted summaries from Paper Search 2.0 save time, allowing researchers to move forward more efficiently. The integrated tools make literature reviews more streamlined and less time-consuming. Future enhancements include expanding databases beyond Semantic Scholar, offering more refined search capabilities, and integrating advanced AI features for deeper insights.
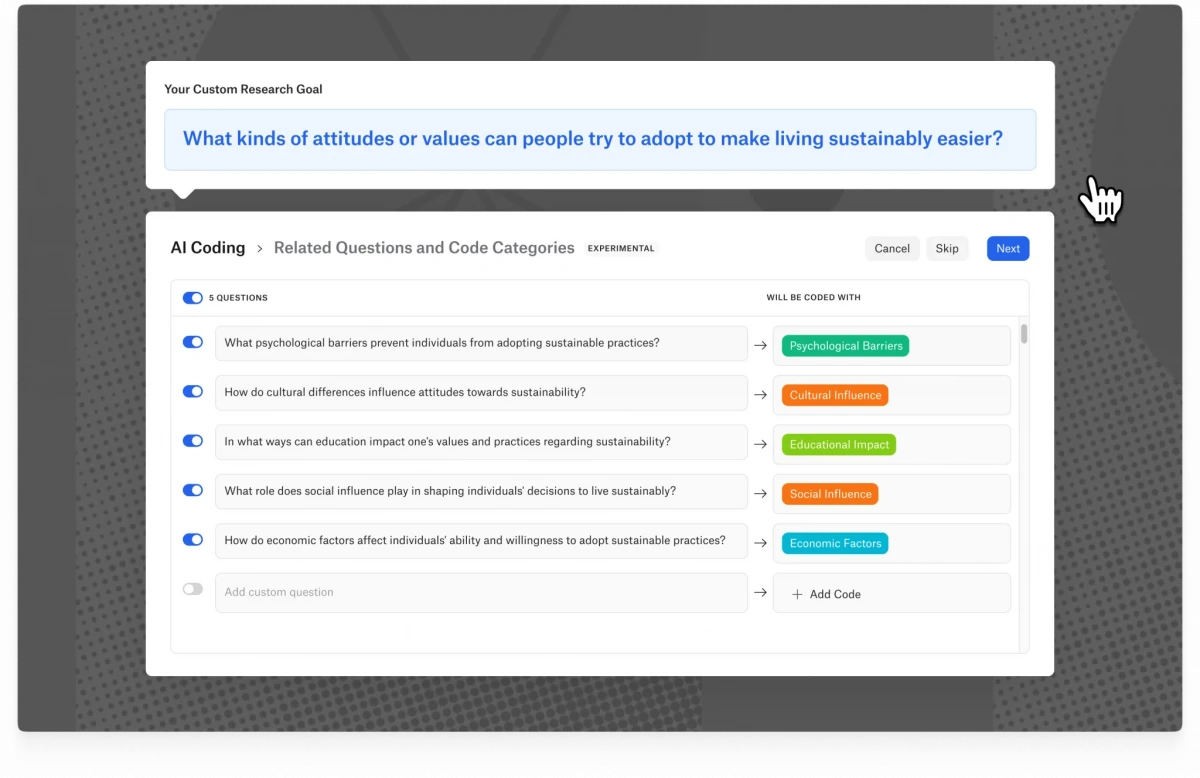
Intentional AI Coding in Desktop and Web
ATLAS.ti's Intentional AI Coding leverages the advanced capabilities of OpenAI's ChatGPT technology. This innovative solution enables researchers to steer automated coding by providing detailed context about their intentions, concepts of interest, and research scope. By doing so, it generates more accurate and relevant coding suggestions tailored to specific research goals. Intentional AI Coding also upholds scientific rigor by creating traceable research questions that link back to the user's methodology and objectives, ensuring transparency throughout the research process.
Moreover, this feature puts the user in full control, allowing them to review, refine, and customize the research questions and code categories before applying them, thereby ensuring the final codes are entirely user-generated. This level of control helps researchers maintain their unique analytical perspectives. Additionally, Intentional AI Coding enhances the speed and depth of insights by improving the precision of coding from the outset, enabling researchers to identify themes and draw robust conclusions more quickly.
In response to privacy concerns, ATLAS.ti has introduced an AI Privacy Mode, allowing users to deactivate OpenAI integration to ensure no data is submitted externally. This feature can also be managed at an organizational level for multi-user licenses, providing consistent privacy standards across all users. Furthermore, the enhanced Conversational AI in ATLAS.ti Web now supports interactions with multiple documents simultaneously, allowing users to engage in natural conversations to explore and compare data across various sources, broadening their analytical scope and facilitating deeper insights. Currently in beta, these advancements promise to significantly enhance the efficiency and effectiveness of qualitative research.
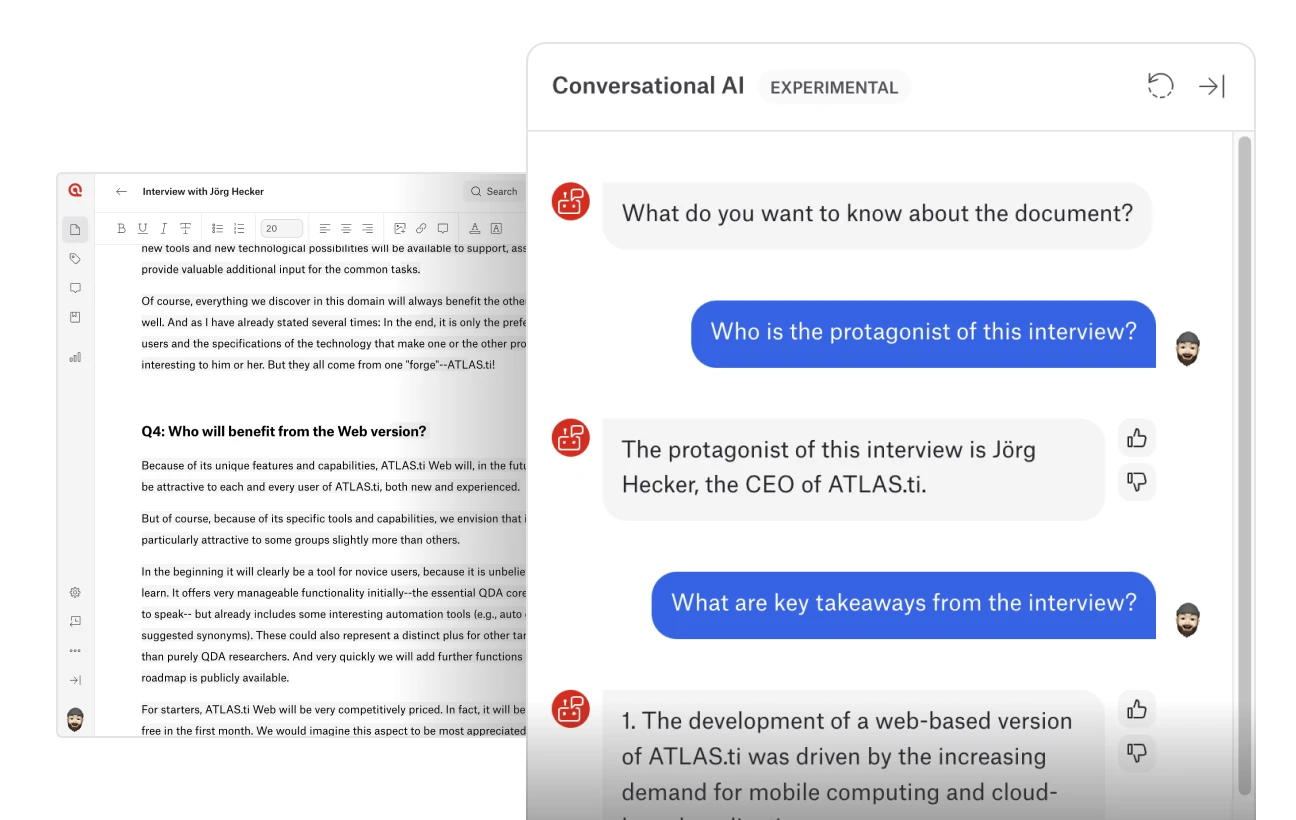
Conversational AI in ATLAS.ti Web
ATLAS.ti has a revolutionary way to interact with your data through natural conversation with its Conversational AI now available for ATLAS.ti Web. This experimental feature leverages OpenAI's ChatGPT technology to transform the research process by allowing users to engage in real-time, natural language dialogues with their documents. You can ask questions, seek clarifications, and navigate your data effortlessly, enabling a more intuitive and efficient way to extract and understand vital information.
The key benefits of this feature include natural interaction, where you converse with your data just as you would with a research partner, and focused navigation, which allows you to locate specific passages and details without manual searching. Enhanced understanding is achieved by clarifying complex passages directly within the chatbot interface, ensuring accurate data interpretation. The user-friendly design of the chatbot makes it easy and accessible, providing real-time assistance to handle extensive documents and reveal insights, connections, themes, and patterns that might otherwise be overlooked.
Users can rate chatbot responses to help train the model, continuously improving its accuracy and relevance. While Conversational AI is currently in an experimental phase and may sometimes produce inaccurate or slower responses, ATLAS.ti is dedicated to refining its performance. This initial version of Conversational AI represents the first step in ATLAS.ti's vision to streamline qualitative data analysis with advanced AI interactions, aiming to create interfaces that simulate collaboration with a knowledgeable human partner. Stay tuned as ATLAS.ti continues to push the boundaries of AI-assisted qualitative research, promising more groundbreaking innovations in the future.
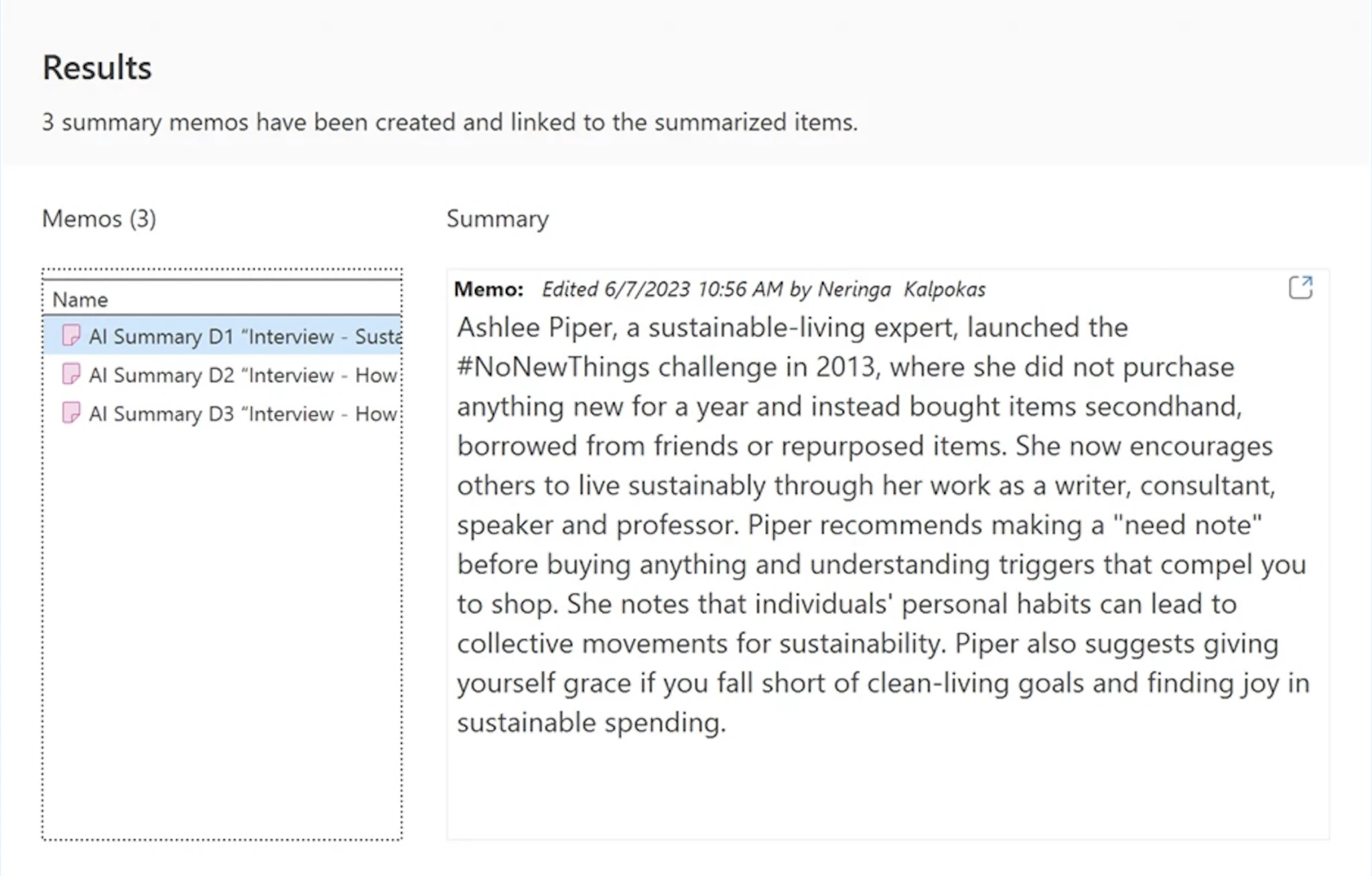
AI Summaries in ATLAS.ti Desktop
ATLAS.ti Desktop AI Summaries is a feature designed to streamline your literature review and qualitative research by providing concise snapshots of large documents and code sets. AI Summaries enhance efficiency by condensing complex information, making it easier to grasp main points swiftly. Integrating seamlessly into memos, the tool supports versatile linking, document conversion, and coding. With a focus on privacy and security, AI Summaries ensure your research data is protected while revolutionizing your research process .
AI future research directions are centred around enhancing the capabilities and usability of literature review tools. Incorporating state-of-the-art NLP technologies, like Large Language Models (LLMs), can significantly improve the performance of these tools, making them more effective and reliable. There is a need to develop advanced interpretability techniques to build trust and provide deeper insights into AI models' decision-making processes. Integrating semantic technologies and creating robust benchmarks and evaluation frameworks will allow for more objective comparisons and assessments of different literature review tools. By addressing these challenges, future AI-enhanced review tools can become more user-friendly and widely adopted, ultimately transforming the systematic review process and contributing to more efficient and comprehensive research methodologies.
ATLAS.ti's AI Lab is continuously working to optimize its AI tools. It maintains transparency by allowing users to view original data behind AI-generated insights which ensures rigorous and precise analysis. Its AI Lab has developed unique tools that extend beyond basic AI functionalities, emphasizing user control and data residency options to address privacy concerns.

Wagner, G., Lukyanenko, R., & Paré, G. (2021). Artificial intelligence and the conduct of literature reviews. Journal of Information Technology, 37(2), 209-226. https://doi.org/10.1177/02683962211048201
Bolaños, F., Salatino, A., Osborne, F., & Motta, E. (2024). Artificial intelligence for literature reviews: Opportunities and challenges. arXiv. https://arxiv.org/abs/2402.08565

Develop powerful literature reviews with ATLAS.ti
Use our intuitive data analysis platform to make the most of your literature review. Get started with a free trial.
RAxter is now Enago Read! Enjoy the same licensing and pricing with enhanced capabilities. No action required for existing customers.
AI-Powered Literature Review: Faster, Deeper, Smarter
Enago Read simplifies literature reviews by delivering smart AI-driven summaries, key insights, real-time discovery, and a copilot that empowers you to master the literature with superhuman speed.
From Surface-Level Exploration to Critical Reading - All in one Place!
Absorb complex research in seconds with intelligent, high-impact summaries and key insights.
AI-powered literature discovery helps you explore related papers and uncover new insights with intelligent recommendations.
Ask questions and receive real-time answers for deeper understanding of complex literature, guiding you through even the most intricate studies.
Literature Review Made Easy
Copilot: go from overwhelmed to enlightened in moments.
Get instant insights, clarity, and deeper understanding in an instant.
Summaries: Decide What to Read, Faster
Reduce information overload by focusing only on papers that matter.
Key Insights: Unlock the Paper's Core Ideas
Quickly understand key takeaways and focus on the most important points of literature.
Discover Related Literature from a 200 Million+ Database
Effortlessly discover literature related to paper you are reading, without getting lost in the endless maze of papers.
Find papers from across the world's largest repositories

Testimonials
Privacy and security of your research data are integral to our mission..

Everything you add or create on Enago Read is private by default. It is visible only to you.

You can put Creative Commons license on original drafts to protect your IP.

We use advanced security protocols including MD5 Encryption, SSL, and HTTPS to secure your data.

Revolutionize Your Research with Jenni AI
Literature Review Generator
Welcome to Jenni AI, the ultimate tool for researchers and students. Our AI Literature Review Generator is designed to assist you in creating comprehensive, high-quality literature reviews, enhancing your academic and research endeavors. Say goodbye to writer's block and hello to seamless, efficient literature review creation.

Loved by over 3 million academics

Endorsed by Academics from Leading Institutions
Join the Community of Scholars Who Trust Jenni AI

Elevate Your Research Toolkit
Discover the Game-Changing Features of Jenni AI for Literature Reviews
Advanced AI Algorithms
Jenni AI utilizes cutting-edge AI technology to analyze and suggest relevant literature, helping you stay on top of current research trends.
Get started

Idea Generation
Overcome writer's block with AI-generated prompts and ideas that align with your research topic, helping to expand and deepen your review.
Citation Assistance
Get help with proper citation formats to maintain academic integrity and attribute sources correctly.

Our Pledge to Academic Integrity
At Jenni AI, we are deeply committed to the principles of academic integrity. We understand the importance of honesty, transparency, and ethical conduct in the academic community. Our tool is designed not just to assist in your research, but to do so in a way that respects and upholds these fundamental values.
How it Works
Start by creating your account on Jenni AI. The sign-up process is quick and user-friendly.
Define Your Research Scope
Enter the topic of your literature review to guide Jenni AI’s focus.
Citation Guidance
Receive assistance in citing sources correctly, maintaining the academic standard.
Easy Export
Export your literature review to LaTeX, HTML, or .docx formats
Interact with AI-Powered Suggestions
Use Jenni AI’s suggestions to structure your literature review, organizing it into coherent sections.
What Our Users Say
Discover how Jenni AI has made a difference in the lives of academics just like you

I thought AI writing was useless. Then I found Jenni AI, the AI-powered assistant for academic writing. It turned out to be much more advanced than I ever could have imagined. Jenni AI = ChatGPT x 10.

Charlie Cuddy
@sonofgorkhali
Love this use of AI to assist with, not replace, writing! Keep crushing it @Davidjpark96 💪

Waqar Younas, PhD
@waqaryofficial
4/9 Jenni AI's Outline Builder is a game-changer for organizing your thoughts and structuring your content. Create detailed outlines effortlessly, ensuring your writing is clear and coherent. #OutlineBuilder #WritingTools #JenniAI

I started with Jenni-who & Jenni-what. But now I can't write without Jenni. I love Jenni AI and am amazed to see how far Jenni has come. Kudos to http://Jenni.AI team.

Jenni is perfect for writing research docs, SOPs, study projects presentations 👌🏽

Stéphane Prud'homme
http://jenni.ai is awesome and super useful! thanks to @Davidjpark96 and @whoisjenniai fyi @Phd_jeu @DoctoralStories @WriteThatPhD
Frequently asked questions
What exactly does jenni ai do, is jenni ai suitable for all academic disciplines, is there a trial period or a free version available.
How does Jenni AI help with writer's block?
Can Jenni AI write my literature review for me?
How often is the literature database updated in Jenni AI?
How user-friendly is Jenni AI for those not familiar with AI tools?
Jenni AI: Standing Out From the Competition
In a sea of online proofreaders, Jenni AI stands out. Here’s how we compare to other tools on the market:
Feature Featire
COMPETITORS
Advanced AI-Powered Assistance
Uses state-of-the-art AI technology to provide relevant literature suggestions and structural guidance.
May rely on simpler algorithms, resulting in less dynamic or comprehensive support.
User-Friendly Interface
Designed for ease of use, making it accessible for users with varying levels of tech proficiency.
Interfaces can be complex or less intuitive, posing a challenge for some users.
Transparent and Flexible Pricing
Offers a free trial and clear, flexible pricing plans suitable for different needs.
Pricing structures can be opaque or inflexible, with fewer user options.
Unparalleled Customization
Offers highly personalized suggestions and adapts to your specific research needs over time.
Often provide generic suggestions that may not align closely with individual research topics.
Comprehensive Literature Access
Provides access to a vast and up-to-date range of academic literature, ensuring comprehensive research coverage.
Some may have limited access to current or diverse research materials, restricting the scope of literature reviews.
Ready to Transform Your Research Process?
Don't wait to elevate your research. Sign up for Jenni AI today and discover a smarter, more efficient way to handle your academic literature reviews.
Educational resources and simple solutions for your research journey

Top AI Tools for Literature Review
As researchers, one of the most important steps in conducting a comprehensive study is to perform a literature review. Some of the common challenges researchers face include the overwhelming volume of literature, difficulty in identifying relevant studies, synthesizing findings, and keeping up with the latest research. Fortunately, AI tools for literature reviews are helping researchers overcome these challenges by providing efficient and effective ways to search, filter, and analyze literature.
In this blog post, we’ll explore some of the top AI tools for literature review and how they can help streamline the research process.
Table of Contents
Top five AI tools for literature review
Conducting literature review is a long and arduous task where researchers have to closely go through vast information sources. However, with these five AI literature review tools, sifting large volumes of information easy.
R Discovery
R Discovery is one of the largest scholarly content repositories allowing you access to 250M+ research papers. R Discovery accelerates your research discovery journey, with latest and relevant content in your area of interest. Below are a few benefits of R Discovery tailored to students and researchers to enhance their research finding and reading experience:
- Personalized Research Reading Feeds – R Discovery curates an academic reading library based on the user’s interests. It provides personalized reading recommendations with daily alerts on top papers, ensuring that users are always updated with the latest and most relevant research in their fields.
- Multiple reading lists – You can create and manage separate reading lists for different literature reviews you’re conducting, keeping your research organized.
- Multilingual & Full-Text Audio Features – R Discovery offers audio versions of research abstracts or full-text articles in more than 30+ languages. This feature is particularly beneficial for non-native English speakers, allowing them to listen to or read research papers in their preferred language, thus enhancing accessibility and comprehension
- Smart Research Alerts – The app sends targeted research alerts and notifications to users based on their reading habits and preferences. Users receive no more than three research paper notifications per day to avoid information overload. The notifications include curated lists of top research papers, updates from preferred journals, and alerts on full-text articles available for reading.
- Integration with Reference Managers – The platform offers auto-sync capabilities with reference managers like Zotero and Mendeley, ensuring that users’ libraries are up to date with the papers they save or remove
Mendeley is a comprehensive reference management software that serves as an essential tool for researchers, academics, and students engaged in literature review and research management. Founded in 2007 by PhD students and acquired by Elsevier in 2013, Mendeley has evolved into a multifaceted platform that facilitates the organization, sharing, and discovery of research papers.
Below are the detailed features of this AI tool for literature reviews:
- Reference Management – Mendeley allows users to store, organize, and search all their references from a single library. This feature simplifies the process of managing a vast amount of literature, making it easier to access and cite these references in future research.
- Viewing and Annotation – Users can open PDFs directly within Mendeley’s PDF viewer, where they can add highlights and notes to the document. These annotations are stored in the user’s Mendeley account, and a new PDF file containing all annotations can be exported, facilitating collaborative review and personal notetaking.
- Collaboration and Sharing – Mendeley supports collaboration through the creation of private groups, allowing users to share references and annotated documents with co-workers or research team members. This feature enhances the collaborative aspect of literature review by enabling shared access to key resources.
- Literature Search and Discovery – While a dedicated literature search feature was removed from the latest version of Mendeley, the platform still offers capabilities for discovering relevant research. Users can import references from other sources and utilize Mendeley’s academic social network to find and share literature.
- Citation and Bibliography Generation – Mendeley simplifies the citation process with Mendeley Cite, an add-in for Microsoft Word that automates the generation of citations and bibliographies in various citation styles. This feature significantly reduces the time and effort required to accurately cite sources during a literature review.
Zotero is an open-access, easy-to-use reference management tool designed to assist with the collection, organization, citation, and sharing of research sources. It serves as a personal research assistant for students, researchers, and academics, helping them manage their literature review process efficiently.
- Reference Management – Zotero allows users to collect citations from various sources, including books, articles, media, webpages, and more. It provides a centralized library where users can organize these references into collections, tag them with keywords, and create saved searches that automatically populate with relevant materials.
- PDF Viewing and Annotation – Zotero has a built-in PDF viewer that enables users to highlight text, add sticky notes, and take snapshots of images or charts within PDFs. Annotations made in the PDF viewer are saved within Zotero and can be extracted as notes, making them fully searchable and easier to organize.
- Collaboration and Sharing – Zotero’s Groups feature allows users to collaborate with other Zotero users through shared libraries. Users can create private, public closed, or public open groups to work on collaborative research projects, distribute course materials, or build collaborative bibliographies
- Citation and Bibliography Generation – Zotero integrates with word processors like Microsoft Word, LibreOffice, and Google Docs to insert citations and bibliographies directly into documents. It supports over 10,000 citation styles, allowing users to format their work to match any style guide or publication
Scholarcy
Scholarcy is an innovative tool designed to assist researchers, students, and academics in managing the often-overwhelming task of conducting literature reviews. It leverages artificial intelligence to automate the extraction of key information from academic papers, creating structured summaries that make it easier to evaluate and understand research articles. Below are the detailed features of Scholarcy that are particularly useful for conducting literature reviews:
- Flashcard Summaries – Scholarcy generates interactive summary flashcards from research papers, book chapters, and other documents. These flashcards highlight key information, providing a compact and easy-to-read overview of the text. This feature is perfect for skim reading or getting to the key points of an article before reading it in full.
- Smart Highlighting and Analysis – The tool guides users to important sections of text and helps interpret them through smart highlighting and analysis. Scholarcy identifies factual statements and findings, highlighting them in different colors (blue for factual statements and orange for findings) to facilitate quick understanding of the content.
- Access to Full Text and Cited Papers – Scholarcy provides convenient access to the full text of articles and cited papers. It uses icons to link directly to the full text and to services like Libkey, which provides access via institutional subscriptions. Additionally, clicking on the Scholarcy icon shows the key findings from cited articles, offering a quick overview of the citation context.
- Literature Discovery and Screening – Scholarcy aids in the discovery and screening of new literature. It can source, screen, and store academic literature more efficiently than traditional methods. The Scholarcy browser extension provides a detailed Synopsis and Highlights for a more comprehensive screening than what an abstract alone can offer. This process can take as little as five minutes, allowing readers to absorb the underlying points of the literature quickly
- Reference Management Integration – Scholarcy allows for the export of flashcards to reference management software like Zotero. This integration enables users to see not only the reference of the article in their Zotero library but also the key highlights and structured summaries generated by Scholarcy.
Unpaywall
Offering open access to scholarly articles, Unpaywall is a free database built from over 50,000 publishers and repositories globally. They also leverage open data sources like PubMed Central, DOAJ, Crossref, and DataCite. Major databases including Dimensions, Scopus, and Web of Science have incorporated Unpaywall. For users with Digital Object Identifiers (DOIs), Unpaywall provides various tools to find open access and full-text articles. Data access is available through REST API, R API Wrapper, the Simple Query Tool, or by downloading the entire dataset. Below are the features of this AI tool for literature review
- Simple Query – The Simple Query Tool offered by Unpaywall helps users to determine if there is an open access (OA) version of a list of articles they are interested in. It is particularly useful for users with a list of Digital Object Identifiers (DOIs) for articles and are seeking free, full-text versions of these articles are available through Unpaywall’s extensive database.
- Browser Extension – The Unpaywall browser extension automatically searches for legally available, free versions of scholarly articles as you browse. When you come across a research article online, the extension checks Unpaywall’s database for an open access version. If such a version is available, a green tab appears on the side of your browser, which you can click to access the full text of the article directly. This feature is powered by an index of over 20 million free, legal full-text PDFs, making it a powerful ally in the quest for open access literature.
There are many AI tools for literature review available for your assistance. However, the best tool for you depends on your specific needs. Do you prioritize comprehensive search and full-text access? Easy organization and citation management? Or perhaps AI-powered summaries to grasp key findings quickly?
Evaluate the functionalities of each tool and choose the one that best complements your research workflow. With the right AI tool, your literature review can become a breeze.
R Discovery is a literature search and research reading platform that accelerates your research discovery journey by keeping you updated on the latest, most relevant scholarly content. With 250M+ research articles sourced from trusted aggregators like CrossRef, Unpaywall, PubMed, PubMed Central, Open Alex and top publishing houses like Springer Nature, JAMA, IOP, Taylor & Francis, NEJM, BMJ, Karger, SAGE, Emerald Publishing and more, R Discovery puts a world of research at your fingertips.
Try R Discovery Prime FREE for 1 week or upgrade at just US$72 a year to access premium features that let you listen to research on the go, read in your language, collaborate with peers, auto sync with reference managers, and much more. Choose a simpler, smarter way to find and read research – Download the app and start your free 7-day trial today !
Related Posts

What is Systematic Sampling: Definition, Advantages, Disadvantages, and Examples

What is Correlational Research: Definition, Types, and Examples
Analyze research papers at superhuman speed
Search for research papers, get one sentence abstract summaries, select relevant papers and search for more like them, extract details from papers into an organized table.

Find themes and concepts across many papers
Don't just take our word for it.
.webp)
Tons of features to speed up your research
Upload your own pdfs, orient with a quick summary, view sources for every answer, ask questions to papers, research for the machine intelligence age, pick a plan that's right for you, get in touch, enterprise and institutions, common questions. great answers., how do researchers use elicit.
Over 2 million researchers have used Elicit. Researchers commonly use Elicit to:
- Speed up literature review
- Find papers they couldn’t find elsewhere
- Automate systematic reviews and meta-analyses
- Learn about a new domain
Elicit tends to work best for empirical domains that involve experiments and concrete results. This type of research is common in biomedicine and machine learning.
What is Elicit not a good fit for?
Elicit does not currently answer questions or surface information that is not written about in an academic paper. It tends to work less well for identifying facts (e.g. "How many cars were sold in Malaysia last year?") and in theoretical or non-empirical domains.
What types of data can Elicit search over?
Elicit searches across 125 million academic papers from the Semantic Scholar corpus, which covers all academic disciplines. When you extract data from papers in Elicit, Elicit will use the full text if available or the abstract if not.
How accurate are the answers in Elicit?
A good rule of thumb is to assume that around 90% of the information you see in Elicit is accurate. While we do our best to increase accuracy without skyrocketing costs, it’s very important for you to check the work in Elicit closely. We try to make this easier for you by identifying all of the sources for information generated with language models.
How can you get in contact with the team?
You can email us at [email protected] or post in our Slack community ! We log and incorporate all user comments, and will do our best to reply to every inquiry as soon as possible.
What happens to papers uploaded to Elicit?
When you upload papers to analyze in Elicit, those papers will remain private to you and will not be shared with anyone else.
How accurate is Elicit?
Training our models on specific tasks, searching over academic papers, making it easy to double-check answers, save time, think more. try elicit for free..

Accelerate your dissertation literature review with AI
Become a lateral pioneer.
Get started for free and help craft the future of research.
Early access. No credit card required.
Introduction
Dissertation writing is part of being a graduate student. There are many different ways to organise your research, and several steps to this process . Typically, the literature review is an early chapter in the dissertation, providing an overview of the field of study. It should summarise relevant research papers and other materials in your field, with specific references. To understand how to write a good literature review, we must first understand its purpose. The goals of a literature review are to place your dissertation topic in the context of existing work (this also allows you to acknowledge prior contributions, and avoid accusations of plagiarism), and to set you up to show you are making a new contribution to the field. Since literature review is repetitive, many students find it tedious. While there are some traditional tools and techniques to help, covered below, they tend to be cumbersome and keyword-based. For this reason, we built a better tool for research and literature review, which I describe in the last section. You can see the Lateral tool in action , and how it makes the literature review a lot easier. To sign up to the tool, click here.
1. Different kinds of reading
We can divide the activity of reading for research into three different kinds:
- Exploratory reading, mostly done in the initial phase;
- Deep reading of highly informative sources; and
- Broad, targeted skim reading of large collections of books and articles, in order to find specific kinds of information you already know exist.
1.1. Exploratory reading
Initially, a research student will need to read widely in a new field to gain fundamental understanding. In this early stage, the goal is to explore and digest the main ideas in existing research. Traditionally, this phase has been a manual process, but there is a new generation of digital tools to aid in getting a quick overview of your field, and more generally to organise your research . This stage can happen both before and after the research topic or question has been formulated. It is often unstructured and full of serendipitous (“happy accidental”) discovery — the student’s job is to absorb what they find, rather than to conduct a targeted search for particular information.
Put another way: You don’t know what you’re looking for ahead of time. By the end of this phase, you should be able to sketch a rough map of your field of study.
1.2. Narrow, deep reading
After the exploratory reading phase, you will be able to prioritise the information you read. Now comes the second phase: Deep, reflective reading. In this phase, your focus will narrow to a small number of highly relevant sources — perhaps one or two books, or a handful of articles — which you will read carefully, with the goal of fully understanding important concepts. This is a deliberative style of reading, often accompanied by reflective pauses and significant note taking. If the goal in the first phase was sketching a map of the globe, the goal in this second phase is to decide which cities interest you most, and map them out in colour and detail.
1.3. Broad, targeted reading
You have now sketched a map of your field of study (exploratory reading), and filled in some parts of this map in more detail (narrow, deep reading). I will assume that by this point, you have found a thesis question or research topic, either on your own, or with the help of an advisor. This is often where the literature review begins in earnest. In order to coherently summarise the state of your field, you must review the literature once again, but this time in a more targeted way: You are searching for particular pieces of information that either illustrate existing work, or demonstrate a need for the new approach you will take in your dissertation. For example,
- You want to find all “methodology” sections in a group of academic articles, and filter for those that have certain key concepts;
- You want to find all paragraphs that discuss product-market fit, inside a group of academic articles.
To return to the map analogy: This is like sketching in the important roads between your favourite cities — you are showing connections between the most important concepts in your field, through targeted information search.

2. Drawbacks of broad targeted reading
The third phase — broad, targeted reading, where you know what kind of information you’re looking for and simply wish to scan a collection of articles or books to find it — is often the most mechanical and time consuming one. Since human brains tend to lose focus in the face of dull repetition, this is also a tedious and error-prone phase for many people. What if you miss something important because you’re on autopilot? Often, students end up speed- or skim reading through large volumes of information to complete the literature review as quickly as possible. With focus and training, this manual approach can be efficient and effective, but it can also mean reduced attention to detail and missed opportunities to discover relevant information. Only half paying attention during this phase can also lead to accidental plagiarism, otherwise known as cryptomnesia: Your brain subconsciously stores a distinctive idea or quote from the existing literature without consciously attributing it to its source reference. Afterwards, you end up falsely, but sincerely believing you created the idea independently, exposing yourself to plagiarism accusations.
3. Existing solutions to speed up literature reviews
Given the drawbacks of manual speed- or skim-reading in the broad reading phase, it’s natural to turn to computer-driven solutions. One popular option is to systematically create a list of search term keywords or key phrases, which can then be combined using boolean operators to broaden results. For example, in researching a study about teenage obesity, one might use the query:
- “BMI” or “obesity” and “adolescents” and not “geriatric”,
to filter for obesity-related articles that do mention adolescents, but don’t mention older adults.
Constructing such lists can help surface many relevant articles, but there are some disadvantages to this strategy:
- These keyword queries are themselves fiddly and time-consuming to create.
- Often what you want to find is whole “chunks” of text — paragraphs or sections, for example — not just keywords.
- Even once you have finished creating your boolean keyword query list, how do you know you haven’t forgotten to include an important search query?
This last point reflects the fact that keyword searching is “fragile” and error-prone: You can miss results that would be relevant — this is known as getting “false negatives” — because your query uses words that are similar, but not identical to words appearing in one or more articles in the library database. For example, the query “sporting excellence” would not match with an article that mentioned only “high performance athletics”.
4. Lateral — a new solution
To make the process of finding specific information in big collections of documents quicker and easier — for example, in a literature review — search, we created the Lateral app , a new kind of AI-driven interface to help you organise, search through and save supporting quotes and information from collections of articles. Using techniques from natural language processing, it understands, out-of-the-box, not only that “sporting excellence” and “high-performance” athletics are very similar phrases, but also that two paragraphs discussing these topics in slightly different language are likely related. Moreover, it also learns to find specific blocks of information, given only a few examples. Want to find all “methodology” sections in a group of articles? Check. How about all paragraphs that mention pharmaceutical applications? We have you covered. If you’re interested, you can sign up today .
5. Final note — novel research alongside the literature review
Some students, to be more efficient, use the literature review process to collect data not just to summarise existing work, but also to support one or more novel theses contained in their research topic. After all, you are reading the literature anyway, so why not take the opportunity to note, for example, relevant facts, quotes and supporting evidence for your thesis? Because Lateral is designed to learn from whatever kind of information you’re seeking, this process also fits naturally into the software’s workflow.
References:
- Is your brain asleep on the job?: https://www.psychologytoday.com/us/blog/prime-your-gray-cells/201107/is-your-brain-asleep-the-job
- Tim Feriss speed reading: https://www.youtube.com/watch?v=ZwEquW_Yij0
- Five biggest reading mistakes: https://www.timeshighereducation.com/blog/five-biggest-reading-mistakes-and-how-avoid-them
- Skim reading can be bad: https://www.inc.com/jeff-steen/why-summaries-skim-reading-might-be-hurting-your-bottom-line.html
- Cryptomnesia: https://en.wikipedia.org/wiki/Cryptomnesia
- Systematic literature review with boolean keywords: https://libguides.library.cqu.edu.au/c.php?g=842872&p=6024187
Lit review youtube intro: https://www.youtube.com/watch?v=bNIG4qLuhJA
Spread the word

There is a better way than Dropbox and Google Drive to do collaborative research
In this blog, I describe the limitations of Dropbox and Google in the space of research, and propose Lateral as the much needed alternative.

Remote group work and the best student collaboration tools
In this blog, I outline some organisational techniques and the best digital collaborative tools for successful student group work.

6 things to consider and organise before writing your dissertation (and how Lateral can help)
I hope the following six things to consider and organise will make the complex dissertation writing more manageable.
Get into flow.

AI for literature reviews
Let ai assist boost your literature review and analysis, how to use ai assist for your literature review.
- Step one: Identify and import your literature
- Step two: Summarize your documents with AI Assist
- Step three: Determine relevance and sort accordingly
- Step four: Reading and rough coding
- Step five: Confirm your initial codings
- Step six: Refine your code system
- Step seven: Analyze your literature
Literature about literature reviews and analysis
Tuesday, September 19, 2023

As you may have noticed, there is a rapid growth in AI-based tools for all types of software packages. We followed this trend by releasing AI Assist – your virtual research assistant that simplifies your qualitative data analysis. In the following, we will present you the tools and functions of AI Assist and how they can facilitate your literature reviews.
Literature reviews are an important step in the data analysis journey of many research projects, but often it is a time-consuming and arduous affair. Whether you are reviewing literature for writing a meta-analysis or for the background section of your thesis, work with MAXQDA! Besides the classic tools of MAXQDA that can facilitate each phase of your literature review, the new tool AI Assist can boost your literature review and analysis in multiple ways.
Year by year, the number of publications grows in almost every field of research – our insights and knowledge likewise. The drawback is that the number of publications might be too high to keep track of the recent developments in your field of research. Consequently, conducting a proper literature review becomes more and more difficult, and the importance of quickly identifying whether a publication is interesting for your research question constantly increases.
Luckily, MAXQDA’s AI Assist tool is here to help. Among others, it can summarize your documents, text segments, and coded segments. But there is more – based on your coded segments AI Assist can generate subcodes suggestions. In the following, we will present you step-by-step instructions on how to use MAXQDA for your literature review and analysis with a special focus on how AI Assist can support you.
Step one of AI for literature reviews: Identify and import your literature
Despite the fact that MAXQDA and AI Assist can facilitate your literature review and analysis in manifold ways, the best advice is to carefully plan your literature review and analysis. Think about the purpose of your literature review and the questions you want to answer. Develop a search strategy which includes, but is not limited to, deciding on literature databases, search terms, and practical and methodological criteria for selecting high-quality scientific literature. Then start your literature review and analysis by searching the identified databases. Before downloading the PDFs and/or bibliographic information (RIS), briefly scan the search results for relevance by reading the title, keywords and abstract. If you find the publication interesting, download the PDF, and let AI Assist help you determining whether the publication falls within the narrower area of your research question.
MAXQDA’s import tab offers import options dedicated to different data types, such as bibliographic data (in RIS file format) and PDF documents. To import the selected literature, just click on the corresponding button, select the data you want to import, and click okay. Alternatively, you can import data simply by drag-and-dropping the data files from your Windows Explorer/Mac Finder window. If you import full texts and the corresponding bibliographic data, MAXQDA automatically connects the full text to the literature entry with an internal link.

Step two of AI for literature reviews: Summarize your documents with AI Assist
Now that you have imported all publications that might be interesting for your research question, it is time to explore whether they are indeed relevant for your literature review and analysis. Before the release of AI Assist, this step typically took a lot of time as you had to go through each paper individually. With the release of AI Assist, MAXQDA can accelerate this step with AI-generated summaries of your publications. For example, you can create AI-generated summaries either for the entire publication or for each chapter (e.g. Introduction, Methods, Results, and so on) individually and base your decision about a paper’s relevance on these summaries. Each AI-generated summary is stored in a memo that is attached to the underlying document or text segment, respectively.
Summarizing text segments with AI Assist just takes a few clicks. Simply highlight a text segment in the Document Browser and choose AI Assist from the context menu. Adjust the settings to your needs and let the large language model do the work for you. To view and edit the summary, double-click on the yellow memo icon attached to the summarized text passage.
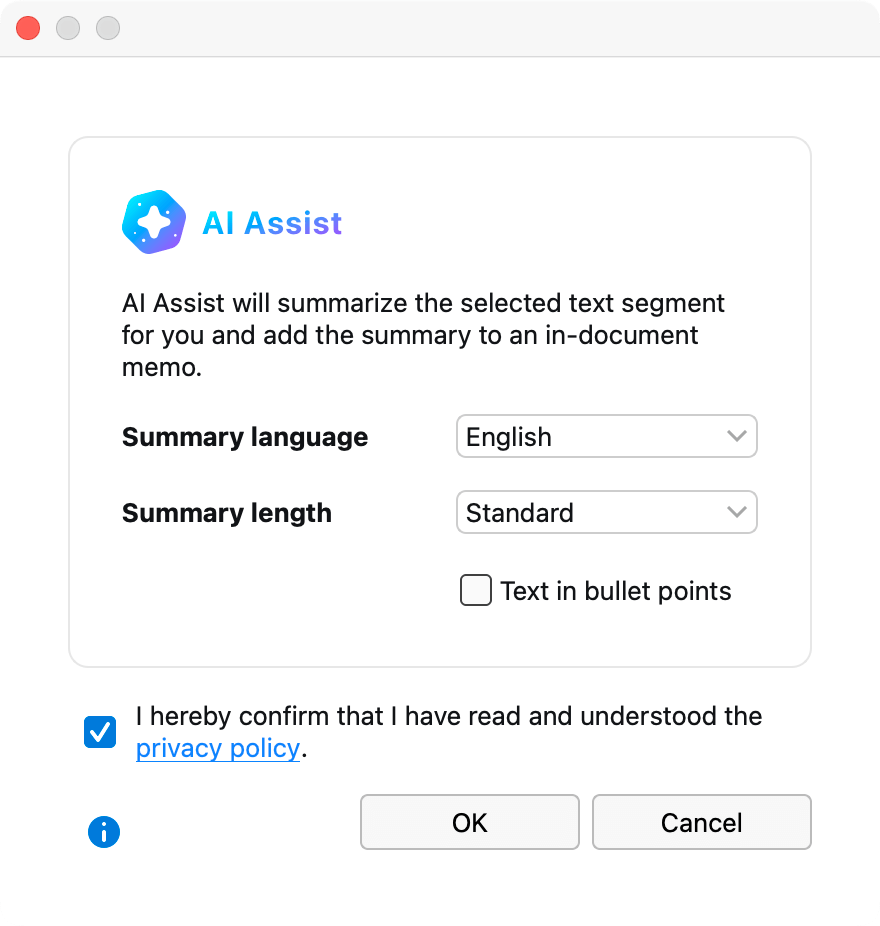
Adjust settings for summarizing text with AI Assist for literature reviews
Step three of AI for literature reviews: Determine relevance and sort accordingly
Instead of reading the entire paper, you can use the AI-generated summaries to determine whether a publication falls within the narrower area of your research question. To do so, it might be helpful to view all memos containing summaries of a specific publication at once. Of course, this is possible with MAXQDA. Go to the Memo tab, click on (In-)document Memos, and click on the publication’s name to view only the AI-generated summaries related to this document. It is important to note that AI-generated summaries are not perfect yet. Therefore, it is advisable to read the entire paper in cases where you have doubts or can’t decide whether the publication is relevant.
Depending on the number of publications in your MAXQDA project, you might want to sort your documents in document groups, for example, based on the relevance for your research question or the topics discussed in the paper. You can easily create a new Document group by clicking on the respective icon in the Document System window. Documents can be added simply via drag-and-drop. Alternatively, you can create Document Sets which are especially helpful when you want to sort your documents by more than one domain (e.g. by relevance and methodology used).

Sort documents in document groups according to their relevance using AI for literature reviews
Step four of AI for literature reviews: Reading and rough coding
Now that you have identified the publications important to your project, it is time to go through the documents. Although, AI Assist can support you at multiple stages of your literature review, it can’t replace the researcher. As a researcher, you still need a deep understanding of your material, analysis methods, and the software you use for analysis. As AI-generated summaries are not perfect yet, you might want to improve the summaries, if necessary, or add information that you consider especially important, e.g. participants’ demographics.
In a next step, it is time to create and apply some codes to the data. A code can be described as a label used to name phenomena in a text or an image. Depending on your approach, you might already have codes in mind (deductive coding) or you plan to generate codes on the basis of the data (inductive coding). No matter your approach – you can use MAXQDA’s advanced tools for coding. In many cases it is best, to start your first round of coding with rather rough codes that you can refine in a later step using the help of AI Assist. You can create codes in the Code System window by clicking on the plus-icon or in the Document Browser by highlighting a text segment via the context menu or the corresponding icons. A code can be applied to the data via drag-and-drop.
Reading and rough coding for AI for literature reviews
Step five of AI for literature reviews: Confirm your initial codings
Though AI Assist can’t validate your codings like a second researcher using intercoder agreement, AI Assist’s Code Summaries can help you to identify whether you have applied the code as intended. The AI-generated Code Summary is a summary of the content of all text segments coded with the corresponing code. This summary might give you an idea of how you have applied the code and if the coded text segments indeed contain what you had in mind when creating the code.
To create a summary of coded segments with AI Assist, simply right-click the code of interest in the Code System and choose AI Assist > Code Summary from the context menu. Adjust language and the summary length to your needs and let AI Assist do the summary for you. As for document summaries, the summary will be stored in a memo which is placed next to the code in the Code System. If the summary doesn’t match your code definition, you might want to review the coded segments and adjust your codings accordingly. By double-clicking on a code, you open the Overview of Coded Segments – a table perfectly suited to go through the coded segments and adjust or remove the codings.
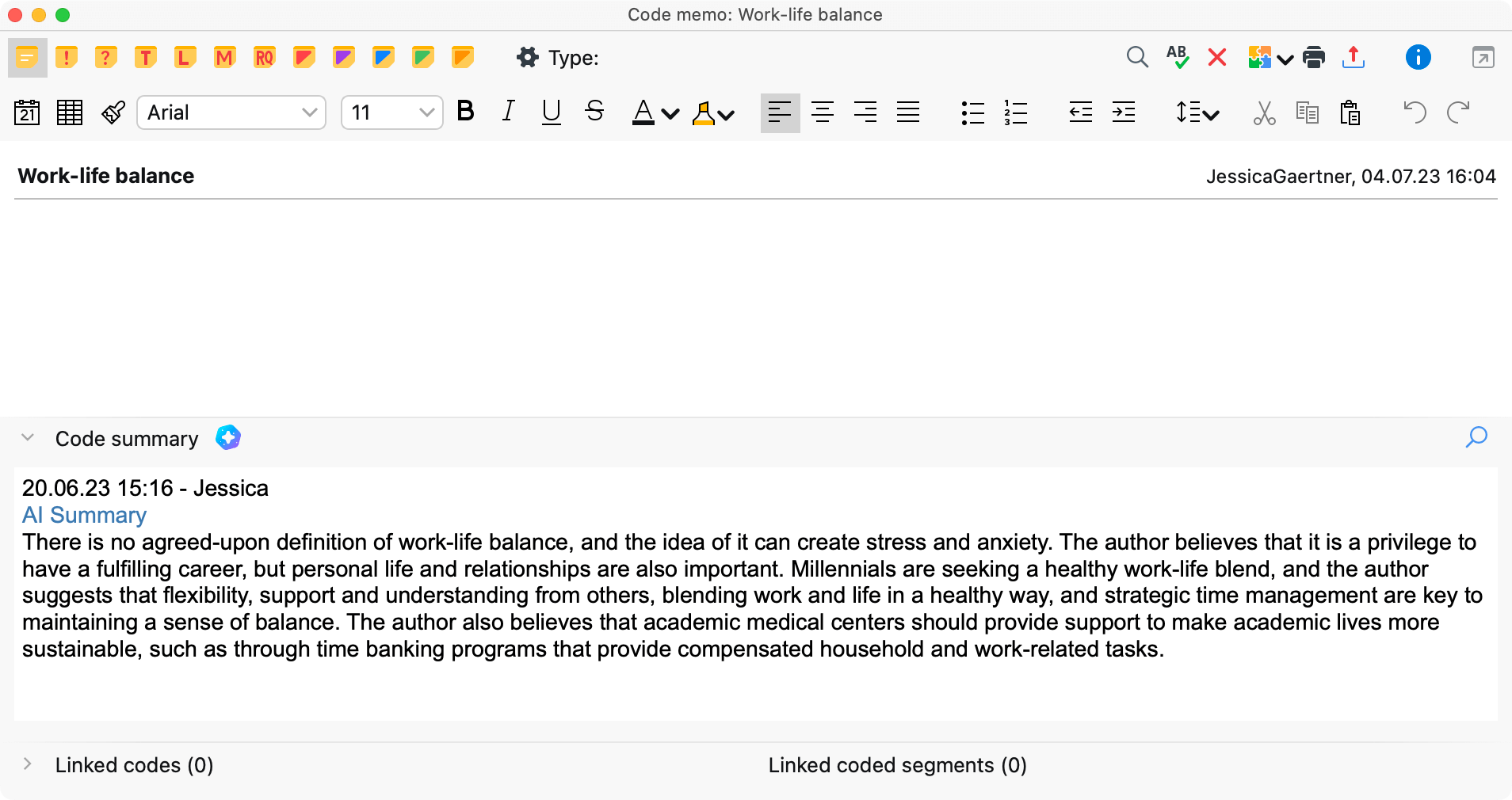
Confirm your initial codings with AI Assist’s Code Summary for literature reviews
Step six of AI for literature reviews: Refine your code system
In case you have applied rather rough codes to your data, your code definitions are probably too broad for you to make sense of the data. Depending on your goals, you might wish to refine these rather broad codes into more precise sub-codes. Again, you can use AI Assist’s power to support this step of your literature review. AI Assist analyzes the text and suggests subcodes while leaving the decision on whether you want to create the suggested sub-codes up to you.
To create AI-generated subcode suggestions, open the context menu of a code and choose AI Assist > Suggest Subcodes. Besides selecting a language, you can ask AI Assist to include examples for each subcode as a bullet list. Like the AI-generated summaries, the code suggestions are stored in the code’s memo. If you are satisfied with the code suggestions, you can create and apply them to your data. Alternatively, you can use the AI-generated code suggestions to confirm the subcodes that you have created.

Use AI Assist’s Suggest Subcodes function to refine your code system for your literature reviews
Step seven of AI for literature reviews: Analyze your literature
Now that you have coded your literature, it’s time to analyze the material with MAXQDA. Although you can use plenty of MAXQDA’s tools and functions even when the material is not coded, other tools require coded segments to be applicable. MAXQDA offers plenty of tools for qualitative data analysis, impossible to mention all. Among others, MAXQDA’s Overview and Summary Tables are useful for aggregating your data. With MAXQDA Visualization Tools you can quickly and easily create stunning visualizations of your data, and with MAXQDA’s Questions-Themes-Theories tool you have a place to synthesize your results and write up a literature review or report.
You can find more information and ideas for conducting a literature review with MAXQDA, here:
Learn more about literature reviews
For information about AI Assist and how to Activate AI Assist, visit:
Learn more about AI Assist
We offer a variety of free learning materials to help you get started with your literature review. Check out our Getting Started Guide to get a quick overview of MAXQDA and step-by-step instructions on setting up your software and creating your first project with your brand new QDA software. In addition, the free Literature Reviews Guide explains how to conduct a literature review with MAXQDA in more detail.

Getting Started with MAXQDA

Literature Reviews with MAXQDA
MAXQDA Newsletter
Our research and analysis tips, straight to your inbox.
- By submitting the form I accept the Privacy Policy.
- FIU Libraries
- Artificial Intelligence Now: ChatGPT + AI Literacy Toolbox
- Literature Reviews with Prompts
Artificial Intelligence Now: ChatGPT + AI Literacy Toolbox: Literature Reviews with Prompts
- Literacy Tutorials
- AI LibGuides
- AI Literacy
- AI + Art News
- AI Plagiarism + Citation
- Which AI tools should I choose?
- Faculty + Academic Writing
- ChatGPT on InfoLit
- Generative AI in the Library
- Art + Image Prompts
- Prompt Literacy
- Text Prompts
- Scholarship + Publishing
- Students + AI Use
- FIU Library Resources
- How does ChatGPT work?
- Zoom Recordings: AI Workshops
- ALA 2024: Breaking Boundaries
Resources on Lit Reviews & AI
- How to Use ChatGPT to Accelerate Literature Review I’d like to show you how I use ChatGPT to speed up my literature review process. The topic I am exploring is heterogeneity(diversity) learning. I started with the keyword “Info-GAIL” as I read about it when using GAIL in the past.
- ChatGPT Simple Literature Review Template The evolution of library services in the digital age has seen a significant shift towards automation and artificial intelligence applications, with OpenAI's ChatGPT being one of the most popular tools. This literature review explores the trends in the application of ChatGPT in library settings, focusing on user engagement and support services from 2015 to 2023.
- ChatGPT as a Tool for Library Research – Some Notes and Suggestions I see ChatGPT and its alternatives as having partial value as tools for library searching. You can use them without any training, but they will perform better when you know some details about them.
9 Ways To Use ChatGPT To Write A Literature Review (WITHOUT Plagiarism) Video
- Step-by-Step
How to Use ChatGPT to Write a Literature Review With Prompts
Dr. Somasundaram R | https://www.ilovephd.com/ | Copyright © 2019-2023 – iLovePhD | May 19, 2023
Writing a literature review can be a challenging task for researchers and students alike. It requires a comprehensive understanding of the existing body of research on a particular topic. However, with the advent of advanced language models like ChatGPT, the process has become more accessible and efficient.
Discover how to effectively utilize ChatGPT as a research assistant to write a comprehensive and SEO-friendly literature review. Follow our step-by-step guide to leverage this powerful tool, optimize your review for search engines, and contribute to the scholarly conversation in your field.
A Step-by-Step Guide: How to Use ChatGPT for Writing a Literature Review
Step 1: Defining Your Research Objective Before diving into the literature review process, it is crucial to define your research objective.
Clearly articulate the topic, research question, or hypothesis you aim to address through your literature review. This step will help you maintain focus and guide your search for relevant sources.
Step 2: Identifying Keywords and Search Terms To effectively use ChatGPT to assist in your literature review, you need to identify relevant keywords and search terms related to your research topic.
These keywords will help you narrow down your search and gather pertinent information. Consider using tools like Google Keyword Planner or other keyword research tools to discover commonly used terms in your field.
Step 3: Familiarizing Yourself with ChatGPT Before engaging with ChatGPT, it is essential to understand its capabilities and limitations. Familiarize yourself with the prompts and commands that work best with the model.
Keep in mind that ChatGPT is an AI language model trained on a vast amount of data, so it can provide valuable insights and suggestions, but it’s important to critically evaluate and validate the information it generates.
Step 4: Generating an Initial Literature Review Outline Start by creating an outline for your literature review. Outline the main sections, such as the introduction, methodology, results, discussion, and conclusion.
Within each section, jot down the key points or subtopics you want to cover. This will help you organize your thoughts and structure your review effectively.
Step 5: Engaging with ChatGPT for Research Assistance Once you have your outline ready, engage with ChatGPT for research assistance.
Begin by providing a clear and concise prompt that specifies the topic, context, and any specific questions you have. For example, “What are the current trends in [your research topic]?” or “Can you provide an overview of the main theories on [your research question]?”
Step 6: Reviewing and Selecting Generated Content ChatGPT will generate a response based on your prompt. Carefully review the content generated, considering its relevance, accuracy, and coherence.
Extract key points, relevant references, and insightful arguments from the response and incorporate them into your literature review. Be sure to cite and attribute the sources appropriately.
Step 7: Ensuring Coherence and Flow While ChatGPT can provide valuable content, it’s important to ensure the coherence and flow of your literature review.
Use your critical thinking skills to connect the generated content with your research objective and existing knowledge. Rearrange, rephrase, and expand upon the generated text to ensure it aligns with the structure and purpose of your review.
Step 8: Editing and Proofreading Once you have incorporated the generated content into your literature review, thoroughly edit and proofread the document.
Check for grammatical errors, consistency in referencing, and overall clarity. This step is crucial to ensure your literature review is polished and professional.
ChatGPT prompts to Write a Literature Review
Prompts you can use when engaging with ChatGPT for research assistance in writing a literature review:
“Can you provide an overview of the main theories and concepts related to [your research topic]?”
“What are the current trends and developments in [your research field]?”
“Can you suggest some key studies or research papers on [specific aspect of your research topic]?”
“What are the main methodologies used in conducting research on [your research topic]?”
“Can you provide a critical analysis of the existing literature on [your research question]?”
“Are there any gaps or areas of controversy in the literature on [your research topic] that need further exploration?”
“What are the key findings and conclusions from the most recent studies on [your research topic]?”
“Can you suggest some reputable journals or publications explore for relevant literature in [your research field]?”
“What are the different perspectives or schools of thought in the literature on [your research topic]?”
“Can you provide a summary of the historical background and evolution of research on [your research topic]?”
Remember to provide clear and specific instructions in your prompts to guide ChatGPT in generating relevant and accurate content for your literature review.
Using ChatGPT to write a literature review can greatly facilitate the research process. By following a step-by-step approach, researchers can effectively leverage ChatGPT’s capabilities to gather insights, generate content, and enhance the quality of their literature review. However, it is important to approach the generated content critically, validate it with reliable sources, and ensure coherence within the review.
- << Previous: Art + Image Prompts
- Next: Prompt Literacy >>
- Last Updated: Oct 24, 2024 1:47 PM
- URL: https://library.fiu.edu/ai
AI-Based Literature Review Tools
- Dialogues: Insightful Facts
- How to Craft Prompts
- Plugins / Extensions for AI-powered Searches
- Cite ChatGPT in APA / MLA
- AI and Plagiarism
- ChatGPT & Higher Education
Selected AI-Based Literature Review Tools
Disclaimer:
- The guide is intended for informational purposes. It is advisable for you to independently evaluate these tools and their methods of use.
- Dimensions (TAMU) - offers several AI-powered features, such as summarization. The free version of Dimensions Research GPT provides access only to open-access publications.
- Statista (TAMU) - Use its Research Assistant to generate insights from queried data..
- News about their AI Assistant (Beta): Web of Science , Scopus , Ebsco , ProQues t, OVID , Dimensions , JStor , Westlaw , and LexisNexis .
Suggestions:
- Please keep these differences in mind when exploring AI-powered academic search engines.

--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------
- https://www.openread.academy/
- Accessed institutionally by Harvard, MIT, University of Oxford, Johns Hopkins, Stanford, and more. ..
- AI-powered Academic Searching + Web Searching - Over 300 million papers and real-time web content.
- Trending and Topics - Browse them to find the latest hot papers. Use Topic to select specific fields and then see their trending.
- Each keyword search or AI query generates a synthesis report with citations. To adjust the search results, simply click on the Re-Generate button to refresh the report and the accompanied citations. After that click on Follow-Up Questions to go deeper into a specific area or subject.
- Use Paper Q&A to interact with a text directly. Examples: " What does this paper say about machine translation ?" ; "What is C-1 in Fig.1?"
- When you read a paper, under Basic Information select any of the following tools to get more information: Basic Information > Related Paper Graph> Paper Espresso > Paper Q&A , and > Notes. The Related Paper Graph will present the related studies in a visual map with relevancy indication by percentage.
- Click on Translation to put a text or search results into another language.
- Read or upload a document and let Paper Espresso analyze it for you. It will organize the content into a standard academic report format for easy reference: Background and Context > Research Objectives and Hypotheses > Methodology > Results and Findings > Discussion and Interpretation > Contributions to the field > Structure and Flow > Achievements and Significance , and > Limitations and Future Work.
SEMANTIC SCHOLAR
- SCIENTIFIC LITERATURE SEARCH ENGINE - finding semantically similar research papers.
- " A free, AI-powered research tool for scientific literature." <https://www.semanticscholar.org/>. But login is required in order to use all functions.
- Over 200 millions of papers from all fields of science, the data of which has also served as a wellspring for the development of other AI-driven tools.
The 4000+ results can be sorted by Fields of Study, Date Range, Author, Journals & Conferences
Save the papers in your Library folder. The Research Feeds will recommend similar papers based on the items saved.
Example - SERVQUAL: A multiple-item scale for measuring consumer perceptions of service quality Total Citations: 22,438 [Note: these numbers were gathered when this guide was created] Highly Influential Citations 2,001 Background Citations 6,109 Methods Citations 3,273 Results Citations 385
Semantic Reader "Semantic Reader is an augmented reader with the potential to revolutionize scientific reading by making it more accessible and richly contextual." It "uses artificial intelligence to understand a document’s structure and merge it with the Semantic Scholar’s academic corpus, providing detailed information in context via tooltips and other overlays ." <https://www.semanticscholar.org/product/semantic-reader>.
Skim Papers Faster "Find key points of this paper using automatically highlighted overlays. Available in beta on limited papers for desktop devices only." <https://www.semanticscholar.org/product/semantic-reader>. Press on the pen icon to activate the highlights.
TLDRs (Too Long; Didn't Read) Try this example . Press the pen icon to reveal the highlighted key points . TLDRs "are super-short summaries of the main objective and results of a scientific paper generated using expert background knowledge and the latest GPT-3 style NLP techniques. This new feature is available in beta for nearly 60 million papers in computer science, biology, and medicine..." < https://www.semanticscholar.org/product/tldr>
- AI-POWERED RESEARCH ASSISTANT - finding papers, filtering study types, automating research flow, brainstorming, summarizing and more.
- " Elicit is a research assistant using language models like GPT-3 to automate parts of researchers’ workflows. Currently, the main workflow in Elicit is Literature Review. If you ask a question, Elicit will show relevant papers and summaries of key information about those papers in an easy-to-use table." <https://elicit.org/faq#what-is-elicit.>; Find answers from 175 million papers. FAQS
- Example - How do mental health interventions vary by age group? / Fish oil and depression Results: [Login required] (1) Summary of top 4 papers > Paper #1 - #4 with Title, abstract, citations, DOI, and pdf (2) Table view: Abstract / Interventions / Outcomes measured / Number of participants (3) Relevant studies and citations. (4) Click on Search for Paper Information to find - Metadata about Sources ( SJR etc.) >Population ( age etc.) >Intervention ( duration etc.) > Results ( outcome, limitations etc.) and > Methodology (detailed study design etc.) (5) Export as BIB or CSV
- How to Search / Extract Data / List of Concept Search -Enter a research question >Workflow: Searching > Summarizing 8 papers> A summary of 4 top papers > Final answers. Each result will show its citation counts, DOI, and a full-text link to Semantic Scholar website for more information such as background citations, methods citation, related papers and more. - List of Concepts search - e.g. adult learning motivation . The results will present a list the related concepts. - Extract data from a pdf file - Upload a paper and let Elicit extract data for you.
- Export Results - Various ways to export results.
- How to Cite - Includes the elicit.org URL in the citation, for example: Ought; Elicit: The AI Research Assistant; https://elicit.org; accessed xxxx/xx/xx
CONSENSUS.APP
ACADEMIC SEARCH ENGINE- using AI to find insights in research papers.
"We are a search engine that is designed to accept research questions, find relevant answers within research papers, and synthesize the results using the same language model technology." <https://consensus.app/home/blog/maximize-your-consensus-experience-with-these-best-practices/>
- Example - Does the death penalty reduce the crime? / Fish oil and depression / (1) Extracted & aggregated findings from relevant papers. (2) Results may include AIMS, DESIGN, PARTICIPANTS, FINDINGS or other methodological or report components. (3) Summaries and Full Text
- How to Search Direct questions - Does the death penalty reduce the crime? Relationship between two concepts - Fish oil and depression / Does X cause Y? Open-ended concepts - effects of immigration on local economics Tips and search examples from Consensus' Best Practice
- Synthesize (beta) / Consensus Meter When the AI recognizes certain types of research questions, this functionality may be activated. It will examine a selection of some studies and provide a summary along with a Consensus Meter illustrating their collective agreement. Try this search: Is white rice linked to diabetes? The Consensus Meter reveals the following outcomes after analyzing 10 papers: 70% indicate a positive association, 20% suggest a possible connection, and 10% indicate no link.
Prompt “ write me a paragraph about the impact of climate change on GDP with citations “
CITATIONS IN CONTEXT
Integrated with Research Solutions.
Over 1.2 billion Citation Statements and metadata from over 181 million papers suggested reference.
How does it work? - "scite uses access to full-text articles and its deep learning model to tell you, for a given publication: - how many times it was cited by others - how it was cited by others by displaying the text where the citation happened from each citing paper - whether each citation offers supporting or contrasting evidence of the cited claims in the publication of interest, or simply mention it." <https://help.scite.ai/en-us/article/what-is-scite-1widqmr/>
EXAMPLE of seeing all citations and citation statements in one place
More information: Scite: A smart citation index that displays the context of citations and classifies their intent using deep learning
- GPT3.5 by OpenAI. Knowledge cutoff date is September 2021.
- Input/ Output length - ChatGPT-3.5 allows a maximum token limit of 4096 tokens. According to ChatGPT " On average, a token in English is roughly equivalent to 4 bytes or characters. English words are typically around 5 characters long. This means that, very roughly, you could fit around 800 to 1000 English words within 4096 tokens."
- According to ChatGPT, the generated responses are non-deterministic by default. So if you run the searches again and get slightly or very different results, it's likely due to this factor.
- ChatGPT may find non-existent references.
- According to this study < https://arxiv.org/ftp/arxiv/papers/2304/2304.06794.pdf > "ChatGPT cites the most-cited articles and journals, relying solely on Google Scholar's citation counts" within the field of environmental science.
- See a case of using ChatGPT40 to extract a PDF file below.
- Example - "INTERVIEW WITH CHATGPT" as a Research Method & Teaching Tool Some researchers began to use this approach to obtain their research data. Try this Google Scholar search link "interview with ChatGPT" or see two articles below: (1) Chatting about ChatGPT: how may AI and GPT impact academia and libraries? BD Lund, T Wang - Library Hi Tech News, 2023 (2) An interview with ChatGPT: discussing artificial intelligence in teaching, research, and practice , G Scaringi, M Loche - 2023
Increased risk of sudden infant death syndrome (SIDS) Increased risk of premature birth Increased risk of low birth weight Increased risk of respiratory problems in newborns Increased risk of respiratory problems in infants exposed to secondhand smoke Increased risk of developing asthma and other respiratory illnesses later in life for infants exposed to secondhand smoke [Note : ChatGPT may generate non-existent references or false knowledge. To find out why Large Language Models hallucinate, check out this Wiki article: Hallucination (artificial intelligence) and this blog post - A Gentle Introduction to Hallucinations in Large Language Models by Adrian Tam ]
Infant death Neonatal mortality (referring specifically to deaths within the first 28 days of life) Perinatal mortality (referring to deaths occurring during the period from 22 completed weeks of gestation to 7 completed days after birth) Early childhood mortality (referring to deaths occurring within the first five years of life) Child mortality (referring to deaths occurring before the age of 18) [Press the Regenerate button to get more.]
- Example - RELATED WORDS What are the related words of infant mortality? Neonatal mortality, Post-neonatal mortality, Stillbirths, Low birth weight, Malnutrition, Infectious diseases, Vaccination, Maternal health, Access to healthcare, Poverty, Social inequality, Sanitation, Hygiene, Water quality, Childbirth complications, Congenital abnormalities, Birth defects, Maternal age, Under-five mortality, Child mortality, Perinatal mortality, Preterm birth, Low birth weight, Sudden infant death syndrome (SIDS), Maternal mortality, Postnatal care, Malnutrition, Immunization, Access to healthcare, Clean water and sanitation, Congenital anomalies, Infant health disparities, Infant mortality rate (IMR), Infant survival.
(("Infant Death"[Mesh] OR "Sudden Infant Death Syndrome"[Mesh] OR "Infant Mortality"[Mesh] OR "Perinatal Mortality"[Mesh] OR "Neonatal Mortality"[Mesh] OR "Fetal Mortality"[Mesh] OR "Infant Death" OR "Sudden Infant Death Syndrome" OR "Infant Mortality" OR "Perinatal Mortality" OR "Neonatal Mortality" OR "Fetal Mortality") AND ("Risk Factors"[Mesh] OR "Etiology"[Mesh] OR "Prevention and Control"[Mesh] OR "Interventions"[Mesh] OR "Public Health"[Mesh] OR "Health Disparities"[Mesh] OR "Healthcare Disparities"[Mesh] OR "Poverty"[Mesh] OR "Malnutrition"[Mesh] OR "Infection"[Mesh] OR "Congenital Abnormalities"[Mesh] OR "Prematurity"[Mesh] OR "Low Birth Weight"[Mesh] OR "Maternal Education"[Mesh] OR "Maternal Health"[Mesh] OR "Risk Factors" OR "Etiology" OR "Prevention and Control" OR "Interventions" OR "Public Health" OR "Health Disparities" OR "Healthcare Disparities" OR "Poverty" OR "Malnutrition" OR "Infection" OR "Congenital Abnormalities" OR "Prematurity" OR "Low Birth Weight" OR "Maternal Education" OR "Maternal Health")) Comments and Suggestions: #1 -This search string may not work on all database platforms. For example, Medline (OVID) may not be able to run it, Medline & CINAHL (EBSCO) can run it without any problems while PubMed will run it but it will indicate which MeSH terms are not indexed. See PubMed's search History and Details under Advanced . #2 - This is just one possible combination. You may need to adjust it based on your task and the databases you are using . #3 - It might be easier if you break the task into 3 parts: ask for synonyms > ask for thesauri terms > and ask it to combine them.
Examples - DATABASE RECOMMENDATIONS Heath Science - Which databases to use to find articles on the EFFECTS OF MATERNAL SMOKING ON INFANT DEATH? PubMed: This is a database of biomedical literature from the National Library of Medicine. It includes articles from medical journals and other sources on a wide range of health-related topics. Embase: This is a database of biomedical literature that covers more international and pharmaceutical content than PubMed. It includes articles from medical journals and other sources on a wide range of health-related topics. Cochrane Library : This is a collection of databases that provides systematic reviews and meta-analyses of healthcare interventions, including interventions related to maternal smoking and infant death. Scopus: This is a multidisciplinary database that covers research from a wide range of fields, including health sciences. It includes articles from scientific journals, books, and conference proceedings. Web of Science: This is a database that covers research from a wide range of fields, including health sciences. It includes articles from scientific journals, books, and conference proceedings. CINAHL: This is a database of nursing and allied health literature, including articles on maternal smoking and infant death. [Note: The response above was copied and pasted directly from ChatGPT. No editing was added.] Education -- I am doing a meta-analysis on k-12 STEM education. What databases should I use? ERIC : This is a database of education research and information, including articles from journals, reports, and conference papers. It is a commonly used database for educational research. Education Source : This is a comprehensive database of education research and information, including articles from journals, books, and conference papers. Web of Science: This is a multidisciplinary database that covers research from a wide range of fields, including education. It includes articles from scientific journals, books, and conference proceedings. Scopus: This is another multidisciplinary database that covers research from a wide range of fields, including education. It includes articles from scientific journals, books, and conference proceedings. PsycINFO: This is a database of psychology research, including articles from journals, books, and dissertations. It can be useful for research on STEM education that includes psychological factors. Cochrane Library : This is a collection of databases that provides systematic reviews and meta-analyses of healthcare interventions including interventions related to education. [Author's note: You could also ask for the GREY LITERATURE sources related to the topic above,]
Example: ACT-AS-AN-EXPERT PROMPT Here's an alternative way to consolidate all the questions into a single consultation by using this popular Act-As-An-Expert prompt, aka, system message prompt: I am going to conduct a systematic review on games and mathematics related to K-12 education I want you to act as a systematic review expert . I will ask you some questions related to my research topic and you advise me on how to do it.
[You may ask one question at a time] My first question is "which databases should I use?" My second question is "what are the relevant subject terms, descriptors and key words?" My third question is "how to do data extraction?" ..... , .... Please note that the above prompts are merely for illustrative purposes. Actual questions may contain more specific instructions. As an example, for the first question, could you provide guidance on how to identify relevant publications? Specifically, can you provide a list of the primary databases and grey literature sources related to my topic?
Example - GREY LITERATURE Please show me all the resources that offer grey literature for conducting research in the field of education ERIC (Education Resources Information Center): https://eric.ed.gov/ OpenGrey: http://www.opengrey.eu/ British Education Index: https://www.ebscohost.com/academic/british-education-index WorldCat: https://www.worldcat.org/ ProQuest Dissertations & Theses Global: https://www.proquest.com/products-services/pqdtglobal.html EThOS (Electronic Theses Online Service): https://ethos.bl.uk/ Institutional Repositories of universities and academic institutions (search for specific institution repositories) American Educational Research Association (AERA): https://www.aera.net/ National Education Association (NEA): https://www.nea.org/ Association for Supervision and Curriculum Development (ASCD): http://www.ascd.org/ RAND Corporation: https://www.rand.org/ National Center for Education Research (NCER): https://ies.ed.gov/ncer/ National Center for Education Statistics (NCES): https://nces.ed.gov/ European Educational Research Association (EERA): https://www.eera-ecer.de/ UNESCO International Institute for Educational Planning (IIEP): http://www.iiep.unesco.org/ Organization for Economic Cooperation and Development (OECD): https://www.oecd.org/education/ Education Development Center (EDC): https://www.edc.org/ National Institute for Educational Policy Research (NIER) - Japan: https://www.nier.go.jp/ Australian Council for Educational Research (ACER): https://www.acer.org/ Canadian Education Association (CEA): https://www.cea-ace.ca/ My topic is about stem education. Which grey literature databases should I use? STEM Education Research Centre (SERC) Grey Literature Database National STEM Education Research Repository (NSERR) National Science Digital Library (NSDL) STEM Education Resource Center (SERC) GrayLit Network STEM Education Grey Literature Library
See other Comparison Examples under Bing AI.
Examples - ZERO SHOT, ONE SHOT & FEW SHOT PROMPTING for literature review See the examples on the left panel- How to Fine Tune Prompts
Example- using ChatGPT4o to Extract sections from PDF
I am going to upload a PDF file. Please extract its Research Objectives, Hypotheses,Methodology and findings for me, and then save it into a text file. ChatGPT40 : Yes, you can upload the PDF..... The uploaded document appears to be a paper titled "......" I'll begin by searching for and extracting the relevant sections. Extracting Sections: Research Objectives and Hypotheses, Methodology, Results and Findings Let me start with the extraction. The information has been saved into a text file. You can download it using the link below:Download Extracted_Info.txt
Bing Chat / Copilot
- Bing Chat has become Copilot.
- CONVERSATIONAL LLM 4.0 + SEARCH ENGINE. Connects to the Internet.
- Three conversation modes: More Creative, More Balanced or More Precise.
- Comparison - Prompt: My topic is about psychology. Which grey literature databases should I use? More Precise - PsycExtra, NIH Library and links to library guides, .gov and .org websites More Balanced - PsycExtra and three main research guides that mention psychology grey literature sources. More Creative- PsycExtra, LILACS, and Worldcat + definition of grey literature + suggestions on searching for professional association websites, newsletters, magazines, standards and guidelines, and other sources that may contain relevant information for your topic.
- Some examples below were tested in Bing Chat only.
- Example - factors of infant mortality Infant mortality is a term that refers to the death of infants before their first birthday. The leading causes of infant mortality include birth asphyxia, pneumonia, congenital malformations, term birth complications such as abnormal presentation of the fetus umbilical cord prolapse, or prolonged labor, neonatal infection, diarrhea, malaria, measles, and malnutrition1. Many situational factors lead to infant mortality such as the pregnant woman’s level of education, environmental conditions, political infrastructure, and level of medical support1. In the United States, a primary determinant of infant mortality risk is infant birth weight with lower birth weights increasing the risk of infant mortality. The determinants of low birth weight include socioeconomic, psychological, behavioral and environmental factors2.
- Example: PEER REVIEWED SOURCES I need to find some peer reviewed articles about infant mortality? The answer includes 5 articles. The first one is “Causes of death and infant mortality rates among full-term births in the United States between 2010 and 2012: An observational study” by Neha Bairoliya and Günther Fink1. A click on " Tell me more about the first article " leads to this summary: " The first article is titled “Causes of death and infant mortality rates among full-term births in the United States between 2010 and 2012: An observational study” and was published in PLOS Medicine on March 20, 2018. The authors are Neha Bairoliya and Günther Fink. The study used linked birth and death records for the period 2010–2012 to identify the state- and cause-specific burden of infant mortality among full-term infants (born at 37–42 weeks of gestation) in the United States. The researchers found that the full-term infant mortality rate (FTIMR) was 2.2 per 1,000 live births overall and ranged between 1.29 (Connecticut) and 3.77 (Mississippi) at the state level. Sudden unexpected death in infancy (SUDI) accounted for 43% of FTIM; congenital malformations and perinatal conditions accounted for 31% and 11.3% of FTIM, respectively. The largest mortality differentials between states with good and states with poor FTIMR were found for SUDI, with particularly large risk differentials for deaths due to sudden infant death syndrome (SIDS) and suffocation." More searches: Please find five empirical studies focusing on the factors influencing adult learners engaged in online education. Please locate five scholarly articles on stress management among college students site:pubmed.ncbi.nlm.nih.gov

The ERIC Thesaurus contains nearly 12,000 terms, including more than 4,500 descriptors and 7,000 synonyms1. You can use the term “Academic Achievement” or its synonyms such as “Educational Achievement”, “Student Achievement”, “Academic Performance”, “Academic Success”, “Learning Achievement” and more. [ Note : The follow-up responses include some additional sources. See two below: https://www.powerthesaurus.org/academic_achievement https://eric.ed.gov/?ti=Achievement+Gap including https://eric.ed.gov/?ti=Academic+Achievement |
My topic is about psychology. Which grey literature databases should I use?
ChatGPT PsycEXTRA : This database is produced by the American Psychological Association (APA) and includes grey literature such as conference papers, white papers, and government reports. OpenGrey : This database provides access to grey literature produced in Europe, including reports, theses, and conference papers Grey Literature Report : This database is produced by The New York Academy of Medicine and includes reports, conference papers, and other grey literature on health and biomedical topics, including psychology. ProQuest Dissertations and Theses Global : This database provides access to graduate-level research, including theses and dissertations related to psychology. Google Scholar: While not exclusively a grey literature database, Google Scholar can be a useful tool for finding conference papers and other grey literature related to psychology. National Technical Information Service (NTIS) : This database provides access to technical reports produced by U.S. government agencies, including those related to psychology. Bing AI - [asked in the Precise mode] For psychology, some recommended grey literature databases include PsycEXTRA and PsyArXiv . PsycEXTRA is a database of grey literature material relating to psychology, behavioral sciences and health. PsyArXiv is a psychology archive designed to facilitate rapid dissemination of psychological research. What other models have been developed based on SERVQUAL?
ChatGPT SERVPERF: This is a modified version of SERVQUAL that measures service performance instead of service quality. It was developed by Cronin and Taylor (1992) and is based on the same five dimensions as SERVQUAL. SERVPERF-GAP: This model, developed by Caruana and Ewing (2006), extends the SERVQUAL model to include three additional gaps that can occur when customers evaluate service quality. E-SERVQUAL: This model, developed by Parasuraman et al. (2005), adapts the SERVQUAL scale to measure the quality of online services. SERVCO: This is a modified version of SERVQUAL that includes a sixth dimension, "cost," in addition to the five original dimensions. It was developed by Sureshchandar et al. (2002) to address the importance of cost in service quality evaluation. SERVQ-HC: This is a modified version of SERVQUAL that is specifically designed for evaluating the quality of health care services. It was developed by Cronin et al. (2000) and includes 20 items across five dimensions. Bing AI The Nordic model of service quality2. The hierarchical model of service quality3. The American Customer Satisfaction Index (ACSI)4.
Bing AI Sidebar - CHAT, THINK, & WRITE
- Examples of Using AI sidebar to Recap Google Scholar findings
Chat - If you highlight a study in the search results of Google Scholar, you will be prompted with a question by AI sidebar asking if you want to send the selected text to it. If you choose to do so, you can then select either EXPLAIN or EXPAND to learn more about the chosen article. It's worth noting that EXPAND provides more detailed information. You can then click on any of the follow-up links to delve further into the selected article or topic. If the text of the chosen study is written in a foreign language, the AI sidebar can also provide an explanation of the text in English. Another option for extracting the key points from a selected article is to submit the URL of the PDF. Insights - Use the insights button to get more ideas about the topic. Compose - Tell the Edge sidebar your topic and let its Compose tool to generate a draft for you. Google Scholar Button- See also the Plugins and Extension page for how to perform a combined search using Google, Google Scholar, and ChatGPT .
Example: FINDING SIMILAR ARTICLES Find similar articles like this one - [then paste in the abstract ] Note - Use this prompt with Bing chat since real-time web info is required.
Examples: SUMMARIZE + REFERENCE Use a basic prompt like " Please summarize . ..." or add a REFERENCE TEXT that provides the necessary information about what you want it to focus or how the summary or resolution should look like. The reference could be a quote, an excerpt, a partial sentence, a conversation, an abstract, a code snippet and so on. Formula : Specify a role > assign a task > provide a reference text. Example#1 - Scholarly Journal Quote
STEP 1 Prompt: As a research assistant specializing in renewable energy, your task is to locate comparable research studies based on the provided abstract from an article: "This paper discusses the perspective of renewable energy (wind, solar, wave and biomass) in the making of strategies for a sustainable development. Such strategies typically involve three major technological changes: energy savings on the demand side, efficiency improvements in the energy production, and replacement of fossil fuels by various sources of renewable energy."
Bing Chat then found 3 documents: the ScienceDirect paper where the abstract came from and two other documents from un.org and frontiersin.org respectively.
STEP 2 Prompt: Please synthesize the findings of the three studies provided above.
Bing Chat: "Sure! Here is a synthesis of the findings from the three studies:
All three studies emphasize the importance of renewable energy in achieving sustainable development and mitigating climate change. They all discuss various strategies and actions that can be taken to increase the use of renewable energy sources and reduce reliance on fossil fuels.
Henrik Lund’s paper discusses the perspective of renewable energy (wind, solar, wave and biomass) in the making of strategies for a sustainable development. Such strategies typically involve three major technological changes: energy savings on the demand side, efficiency improvements in the energy production, and .....
The United Nations outlines five critical actions the world needs to prioritize now to transform our energy systems and speed up the shift to renewable energy: Make renewable energy technology a global public good, Improve global access to components and raw materials......
“Sustainable Energy Transition for Renewable and Low Carbon Grid” discusses that a sustainable global electricity transition will entail increased use of renewable energy sources particularly wind and solar, nuclear energy as a low carbon energy source, electrification of transport and.......
In summary , these studies suggest that transitioning to renewable energy sources is crucial for sustainable development and mitigating climate change. This can be achieved through various strategies such as increasing energy efficiency, improving access to renewable technology and materials, leveling the playing field for renewables, ending fossil fuel subsidies, and redirecting investments towards renewables." Example #2 Y ou are an expert researcher, given the following article, please discuss its key findings. Reference: How Large Language Models are Transforming Machine-Paraphrased Plagiarism at https://arxiv.org/abs/2210.03568 This is another way to put REFERENCE TEXT. As a side not, role specifying or the use of persona is important as it can impact the level of responses obtained. Just imagine if the designated role were " You are a high school student... ." Example #3 Please read this article and summarize it f or me - "Who Should I Trust: AI or Myself? Leveraging Human and AI Correctness Likelihood to Promote Appropriate Trust in AI-Assisted Decision-Making" and then find 5 similar studies and please also summarize each for me.

CONNECTED PAPERS
- RELATED STUDIES
- Uses visual graphs or other ways to show relevant studies. The database is connected to the Semantic Scholar Paper Corpus which has compiled hundreds of millions of published papers across many science and social science fields.
- See more details about how it works .
- Example - SERVQUAL and then click on SELECT A PAPER TO BUILD THE GRAPH > The first paper was selected. Results: (1) Origin paper - SERVQUAL: A multiple-item scale for measuring consumer perceptions of service quality + Connected papers with links to Connected Papers / PDF / DOI or Publisher's site / Semantic Scholar / Google Scholar. (2) Graph showing the origin paper + connected papers with links to the major sources . See above. (3) Links to Prior Works and Derivative Works See the detailed citations by Semantic Scholar on the origin SERVQUAL paper on the top of this page within Semantic Scholars.
- How to Search Search by work title. Enter some keywords about a topic.
- Download / Save Download your saved Items in Bib format.
PAPER DIGEST
- SUMMARY & SYNTHESIS
- " Knowledge graph & natural language processing platform tailored for technology domain . <"https://www.paperdigest.org/> Areas covered: technology, biology/health, all sciences areas, business, humanities/ social sciences, patents and grants ...

- LITERATURE REVIEW - https://www.paperdigest.org/review/ Systematic Review - https://www.paperdigest.org/literature-review/
- SEARCH CONSOLE - https://www.paperdigest.org/search/ Conference Digest - NIPS conference papers ... Tech AI Tools: Literature Review | Literature Search | Question Answering | Text Summarization Expert AI Tools: Org AI | Expert search | Executive Search, Reviewer Search, Patent Lawyer Search...
Daily paper digest / Conference papers digest / Best paper digest / Topic tracking. In Account enter the subject areas interested. Daily Digest will upload studies based on your interests.
RESEARCH RABBIT
- CITATION-BASED MAPPING: SIMILAR / EARLY / LATER WORKS
- " 100s of millions of academic articles and covers more than 90%+ of materials that can be found in major databases used by academic institutions (such as Scopus, Web of Science, and others) ." See its FAQs page. Search algorithms were borrowed from NIH and Semantic Scholar.
The default “Untitled Collection” will collect your search histories, based on which Research Rabbit will send you recommendations for three types of related results: Similar Works / Earlier Works / Later Works, viewable in graph such as Network, Timeline, First Authors etc.
Zotero integration: importing and exporting between these two apps.
- Example - SERVQUAL: A multiple-item scale for measuring consumer perceptions of service quality [Login required] Try it to see its Similar Works, Earlier Works and Later Works or other documents.
- Export Results - Findings can be exported in BibTxt, RIS or CSV format.
CITING GENERATIVE AI
- How to cite ChatGPT [APA] - https://apastyle. apa.org/blog /how-to-cite-chatgpt
- How to Cite Generative AI [MLA] https://style. mla.org /citing-generative-ai/
- Citation Guide - Citing ChatGPT and Other Generative AI (University of Queensland, Australia)
- Next: Dialogues: Insightful Facts >>
- Last Updated: Oct 18, 2024 2:10 PM
- URL: https://tamu.libguides.com/c.php?g=1289555
What is the future of human-generated systematic literature reviews in an age of artificial intelligence?
- Published: 30 October 2024
Cite this article

- Joern Block ORCID: orcid.org/0009-0007-7199-6107 1 &
- Andreas Kuckertz 1 , 2
74 Accesses
Explore all metrics
Recent advancements in artificial intelligence (AI) enable the rapid collection and organization of academic research with the push of a button, prompting a reevaluation of the role and value of human-generated systematic literature reviews (SLRs). In this editorial, we explore the value of the human element in producing SLRs, a method originally designed to reduce human bias in literature synthesis. Throughout the three stages of an SLR—design, study collection and selection, and interpretation and communication—we argue that the human element is not just a potential source of bias but also an essential contributor of value. The value of human-generated SLRs lies in (1) formulating the right questions, (2) strategic inefficiency in literature search and selection, and (3) individualized problematizing and theorizing. To better leverage the human element in SLRs, we call for new forms of hybrid SLRs that combine traditional systematic review methods with empirical fieldwork.
Avoid common mistakes on your manuscript.
1 Introduction
Advancements in artificial intelligence (AI), particularly in large language models, are transforming the landscape of academic research. This transformation is especially pronounced with systematic literature reviews (SLRs) that do not involve empirical fieldwork. In this area, we are seeing generative AI for natural language generation being increasingly tied to scientific literature databases with impressive performance (Nicholson et al. 2021 ; Skarlinski et al. 2024 ). Additionally, most of us have probably received emails promoting courses designed for researchers titled, for instance, ‘AI-Powered Systematic Review, ' which promise to teach participants how to leverage AI to automate and enhance every stage of an SLR, from literature search and selection to data synthesis. While such AI-powered SLRs may not yet be perfect, AI has already proven to be able to adapt to systematically collecting, selecting, and synthesizing literature.
Moreover, AI is becoming increasingly capable of interpreting and evaluating the content of identified articles, though biases may persist. The primary goal of SLRs today is to synthesize academic research in a structured and transparent manner, interpret the findings, and identify future research directions (Kuckertz and Block 2021 ). By following a clear and systematic literature selection process, such as the one outlined in the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement (Liberati et al. 2009 ; Moher et al. 2009 ), human subjectivity is significantly reduced, particularly in the selection of studies for the review. Viewing this process as an algorithm is not far-fetched, and we can anticipate AI increasingly surpassing human capabilities in executing such tasks. Mollick’s ( 2024 ) observation that today’s AI tools are the ‘worst’ we will ever work with underscores the potential for future developments, especially considering the remarkable capabilities these tools already demonstrate. Against these backgrounds and given that SLRs are a central focus of Management Review Quarterly , the journal must consider a simple but critical question: What is the future of human-generated SLRs in an age of AI?
This editorial aims to explore this question from the perspective of management research, offering insights and considerations for editors and reviewers evaluating SLRs submitted to the journal. What is clear, from a reader’s perspective, is that SLRs will always be valuable, whether they are generated by humans and published as peer-reviewed papers or generated quickly by a (reliable and unbiased) AI tool. However, as gatekeepers of a journal that focuses on human-generated research, we can only consider articles where the human element is clearly evident and provides substantial value beyond what AI tools can achieve alone.
Therefore, in this editorial, we critically examine the value of traditional human-generated SLRs as they exist today. Building on this analysis, we further explore the future of human-generated SLRs in an evolving landscape. The discussion of the future of human-generated SLRs is organized around the three stages of conducting an SLR: the design stage, the study collection and selection stage, and the interpretation and communication stage. For each stage, we discuss the role and value of human researchers.
2 Design stage: the value of human researchers lies in formulating the right questions
Like any research, conducting an SLR requires a straightforward research question and a solid epistemic interest. Literature reviews have always needed to justify the relevance of their question, especially in light of prior reviews. With AI-supported reviews, this need becomes even more pressing. So, why do we need the human factor to formulate convincing research questions for an SLR?
First, one could take a normative perspective and argue that determining a research question should never be delegated to AI. Conducting an SLR has significant implications, as it shapes future research and sets the agenda for new inquiries. While relying on AI to generate research questions may offer a quick solution for those lacking experience or originality, it comes at the cost of autonomy. Researchers may find themselves pursuing topics they have not chosen independently, which can diminish motivation and result in uninspired work. As well-crafted SLRs can shape the direction of an entire field, this loss of autonomy affects not only individual researchers but the broader academic community as well. In addition to the risk of diminished motivation and uninspired work, there is also the danger that future research agendas may be steered towards areas with limited practical impact. AI, trained on historical data, bases its predictions on past trends and is less capable of accounting for current or emerging developments that could significantly influence management practice.
Second, good research questions arise from a deep engagement with the literature. Effective questions emerge through a dynamic process that involves critically examining and problematizing existing research rather than merely filling gaps in the literature (Alvesson and Sandberg 2020 ). Researchers must challenge current work in light of their own experiences, interests, and identities, i.e., what Shepherd et al. ( 2021 ) essentially called ‘me-search’ instead of ‘re-search.’ Such an approach is essential also for the future of human-generated SLRs. Simply deriving research questions from existing research, a task AI can perform, may no longer be sufficient. Researchers motivating the research questions for their SLR might want to complement their engagement with the literature with some empirical fieldwork. They could step outside their offices to engage directly with the world of management and then write practice-inspired SLRs. By deriving the research questions of an SLR in an abductive way from practical observations and aligning them with existing literature, SLRs can address questions with both academic and practical significance. This interplay between research questions, fieldwork, and literature engagement will always be challenging for AI.
3 Study collection and selection stage: the value of human researchers lies in strategic inefficiency
The data collection phase, that is, identifying and selecting suitable papers for an SLR, is undoubtedly one of the most time-consuming and laborious parts of any such project. It might be tempting to delegate precisely this part of the SLR creation to an AI; the immense time savings that can result from doing so potentially go hand in hand with tremendous efficiency gains. However, given the current state of development of AI, it cannot yet be assumed that it would be better or even just as good as humans at adequately assessing the actual fit of a study to the SLR and its quality of craftsmanship.
Moreover, many valuable insights are at risk of being lost if we focus solely on efficiency. We know from other contexts, especially when it comes to creativity, originality, and quality, that a focus on efficiency alone does not necessarily lead to optimal results. Taking detours and not always following the most direct path to a goal can be valuable. This approach, which we call strategic inefficiency, may not be intentional but can be an inevitable and potentially beneficial part of the process. By allowing room for exploration and flexibility, these detours may lead to unexpected insights, fostering creativity and innovation.
Engaging deeply with a particular literature stream, for example, to determine how a particular study relates to the inclusion and exclusion criteria of an SLR, is not only tedious but also potentially enlightening. Some readers may remember wandering the aisles of a physical library in search of a particular book, uncovering unexpected treasures on the shelves to the left and right of the work they were actually looking for. Granted, those days are long gone, and there is no reason to wish for a return to them, but independently searching digital databases and evaluating individual studies, as well as the unexpected thoughts triggered in the process, are of immense value. At its core, it is about serendipity: whenever people give meaning to unplanned discoveries with a positive effect (Busch 2024 ), great value is potentially created. That also applies to the process of creating an SLR. As an example of strategic inefficiency and building on Fisch and Block’s ( 2018 ) six tips for SLRs, Clark et al. ( 2021 ) emphasize the value of team discussions during literature selection, increasing confidence in the selection criteria and their application. This value generated by social interaction within the research team is challenging to achieve in an AI-generated SLR.
4 Interpretation and communication stage: the value of human researchers lies in individualized problematizing and theorizing
The most uninformative SLRs that Management Review Quarterly regularly rejects are those that reiterate the literature without adding any additional insights to the analysis. To make an SLR both informative and engaging, two key strategies are problematizing existing research or developing theories that go beyond the status quo revealed by the SLR.
In the social sciences, and hence in management research, there are always alternative approaches to problematizing and theorizing. Despite the objective nature of SLRs in data collection and literature selection, our experience shows that two independent author teams tackling the same research question with an SLR will invariably reach different conclusions. This fact is not a limitation of the method; instead, it underscores the importance of the human factor in conducting SLRs.
An AI-generated interpretation would not lead to a definitive solution; instead, it would represent one of many possible answers comparable to the varied results from different research groups exploring the same research question. Consequently, humans cannot rely solely on AI to do the work for them. Each interpretation carries a unique perspective shaped by individual approaches to problematizing and theorizing. Human researchers must articulate their unique interpretative perspective, a meta-level task that is currently challenging for AI to accomplish. Once again, as seen in the study collection and selection stage, social interaction within a research team can lead to a better and more transparent interpretation (Clark et al. 2021 ).
5 Upholding the human factor: developing novel forms of hybrid SLRs that involve empirical fieldwork
We posit that while SLRs strive to minimize subjectivity, the human factor in their preparation remains essential, even as AI approaches the potential of superintelligence—something it is still far from achieving. Human researchers can enhance the value of an SLR at all three stages: design, study collection and selection, and interpretation and communication.
However, it is crucial to clarify that our arguments supporting the human factor should not be interpreted as arguments against AI. Rather than an either/or scenario, we advocate for increased efficiency through the effective combination of human intelligence and AI. AI users increasingly function as “cyborgs” (Dell’Acqua et al. 2023 ), integrating their contributions with those of AI.
This development paves the way for new types of SLRs, and Management Review Quarterly welcomes such innovations. Hybrid SLRs are one potential approach as long as the role of the AI component is made transparent. Such hybrid SLRs could integrate the SLR method with empirical fieldwork, utilizing both quantitative and qualitative methods. Such an empirical element could be, in particular, incorporated during the design and interpretation stages of an SLR to enhance the relevance of the SLR’s results for practical application and to strengthen their robustness against misinterpretation. We invite (human) management researchers to submit such hybrid SLRs to us. Management Review Quarterly will continue to play an active role in developing and adapting the SLR method to ensure its relevance in an age of AI. We are living in exciting times; by leveraging the strengths of both humans and machines, we can continue to use SLRs to generate valuable insights for both management theory and practice.
Alvesson M, Sandberg J (2020) The problematizing review: a counterpoint to Elsbach and Van Knippenberg’s argument for integrative reviews. J Manag Stud 57(6):1290–1304. https://doi.org/10.1111/joms.12582
Article Google Scholar
Busch C (2024) Towards a theory of Serendipity: a systematic review and conceptualization. J Manage Stud 61(3):1110–1151. https://doi.org/10.1111/joms.12890
Clark WR, Clark LA, Raffo DM, Williams RI (2021) Extending Fisch and Block’s (2018) tips for a systematic review in management and business literature. Manage Rev Q 71:215–231
Dell’Acqua F, McFowland III E, Mollick ER, Lifshizt-Assaf H, Kellog K, Rajendran S, Lakhani KR (2023) Navigating the jagged technological frontier: field experimental evidence of the effects of AI on knowledge worker productivity and quality (No. 24–013)
Fisch C, Block J (2018) Six tips for your (systematic) literature review in business and management research. Manage Rev Q 68:103–106
Kuckertz A, Block J (2021) Reviewing systematic literature reviews: ten key questions and criteria for reviewers. Manage Rev Q. https://doi.org/10.1007/s11301-021-00228-7
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JPA, Moher D (2009) The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med. https://doi.org/10.1371/journal.pmed.1000100
Moher D, Liberati A, Tetzlaff J, Altman DG, Group TP (2009) Preferred reporting items for systematic reviews and Meta-analyses: the PRISMA Statement. PLoS Med 6(7):e1000097. https://doi.org/10.1371/journal.pmed.1000097
Mollick E (2024) Co-intelligence. Living and working with AI. Penguin Portfolio
Google Scholar
Nicholson JM, Mordaunt M, Lopez P, Uppala A, Rosati D, Rodrigues NP, Rife SC (2021) Scite: a smart citation index that displays the context of citations and classifies their intent using deep learning. Quant Sci Stud 2(3):882–898. https://doi.org/10.1162/qss_a_00146
Shepherd DA, Wiklund J, Dimov D (2021) Envisioning entrepreneurship’s future: introducing me-search and research agendas. Entrepreneurship Theory Pract 45(5):955–966
Skarlinski MD, Cox S, Laurent JM, Braza JD, Hinks M, Hammerling MJ, Ponnapati M, Rodriques SG, White AD (2024) Language agents achieve superhuman synthesis of scientific knowledge. FutureHouse Inc., San Francisco, CA
Download references
The authors have not disclosed any funding.
Author information
Authors and affiliations.
University of Trier, Trier, Germany
Joern Block & Andreas Kuckertz
Entrepreneurship Research Group, University of Hohenheim, Stuttgart, Germany
Andreas Kuckertz
You can also search for this author in PubMed Google Scholar
Corresponding author
Correspondence to Joern Block .
Ethics declarations
Conflict of interest.
The authors have not disclosed any competing interests.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Reprints and permissions
About this article
Block, J., Kuckertz, A. What is the future of human-generated systematic literature reviews in an age of artificial intelligence?. Manag Rev Q (2024). https://doi.org/10.1007/s11301-024-00471-8
Download citation
Published : 30 October 2024
DOI : https://doi.org/10.1007/s11301-024-00471-8
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Systematic literature review
- Artificial intelligence
- Human factor
JEL Classification
- Find a journal
- Publish with us
- Track your research
This manuscript presents a comprehensive review of the use of Artificial Intelligence (AI) in Systematic Literature Reviews (SLRs). A SLR is a rigorous and organised methodology that assesses and integrates prior research on a given topic. Numerous tools have been developed to assist and partially automate the SLR process. The increasing role of AI in this field shows great potential in providing more effective support for researchers, moving towards the semi-automatic creation of literature reviews. Our study focuses on how AI techniques are applied in the semi-automation of SLRs, specifically in the screening and extraction phases. We examine 21 leading SLR tools using a framework that combines 23 traditional features with 11 AI features. We also analyse 11 recent tools that leverage large language models for searching the literature and assisting academic writing. Finally, the paper discusses current trends in the field, outlines key research challenges, and suggests directions for future research. We highlight three primary research challenges: integrating advanced AI solutions, such as large language models and knowledge graphs, improving usability, and developing a standardised evaluation framework. We also propose best practices to ensure more robust evaluations in terms of performance, usability, and transparency. Overall, this review offers a detailed overview of AI-enhanced SLR tools for researchers and practitioners, providing a foundation for the development of next-generation AI solutions in this field.
Introduction
In this page, we report the tables we have developed when conducting our analysis on the Systematic Literature Review Tools.
Systematic Literature Review Tools analysed through AI and Generic Features
Screening phase of systematic literature review tools analysed through ai features.
- Extraction Phase of Systematic Literature Review Tools analysed through AI Features
- Systematic Literature Review Tools analysed based on AI Features
Figures of the paper
Prisma checklist.
- Codebase Snowballing
How to cite
In this section, we report three tables that describe the 21 systematic literature review tools examined according to both generic and AI-based features. In Section 1.1 and Section 1.2 , we present the analysis of the AI features for the screening and the extraction phases, respectively. In Section 1.3 , we report the analysis of the tools according to the generic features. A copy of these tables is persistently available on ORKG.
-: No information available
Extraction phase of Systematic Literature Review Tools analysed through AI Features
Systematic literature review tools analysed based on general features.
-: No information available NA: Not applicable because the tools are specifically for extraction
In this section we attach all the figures of the mauscript in high defininition (300DPI).

In the following table we report our PRISMA checklist, using the model from "Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 2021;372:n71. doi: 10.1136/bmj.n71".
Here is the codebase we developed for the snowballing search on Semantic Scholar.
F. Bolaños Burgos, A. Salatino, F. Osborne, and E. Motta. Artificial intelligence for systematic literature reviews: Opportunities and challenges. Submitted to Artificial Intelligence Review, 2024.
This work is licensed under CC BY 4.0

Free Literature Review Generator
Create a Well-Structured and Informative Literature Review that Sets the Stage for Your Research
Are you struggling to write a comprehensive literature review that effectively synthesizes the existing research on your topic?
Do you want to create a well-structured and informative literature review that sets the stage for your own research?
Look no further!
Our Literature Review Generator is a free AI writing tool designed to help you streamline your research and create a high-quality literature review.
How it Works
Our Literature Review Generator uses advanced AI technology to analyze your input and generate a comprehensive literature review that is tailored to your specific needs. Simply provide some basic information about your research topic, and our algorithm will provide you with a well-structured and informative literature review that synthesizes the existing research on your topic.
Key Features:
- AI-powered literature review generation : Our algorithm uses natural language processing and machine learning to generate a comprehensive literature review that is tailored to your specific needs.
- Free and easy to use : Our Literature Review Generator is completely free and easy to use, with no registration or subscription required.
Benefits of Using Literature Review Generator
- Streamline your research : Our Literature Review Generator saves you time and effort in researching and writing your literature review, allowing you to focus on other aspects of your research.
- Create a comprehensive literature review : Our algorithm provides you with a comprehensive literature review that synthesizes the existing research on your topic, helping you to identify gaps in the literature and areas for further research.
- Improve your writing : Our Literature Review Generator provides you with a well-structured and informative literature review that sets the stage for your own research, helping you to improve your writing and create a high-quality research paper.
- Enhance your research credibility : A comprehensive literature review is essential for establishing the credibility of your research, and our Literature Review Generator helps you to create a literature review that is thorough, well-structured, and informative.
How to Use:
- Provide some basic information about your research topic, including the topic and scope of your research.
- Our algorithm will generate a comprehensive literature review that synthesizes the existing research on your topic.
- Review and refine the literature review to ensure that it meets your needs and is free of errors.
Example Use Cases:
- Use our Literature Review Generator to create a comprehensive literature review for your research paper, thesis, or dissertation.
- Streamline your research with a well-structured and informative literature review that sets the stage for your own research.
- Improve your academic writing with a literature review that is thorough, well-structured, and informative.
Try Literature Review Generator Now!
Don’t let writer’s block hold you back!
Try our Literature Review Generator today and create a comprehensive literature review that sets the stage for your research.
Try other free writing tools
Acronym generator.
Boost your productivity with our lightning-fast Acronym Generator! Create catchy and memorable acronyms in seconds and take your writing to the next level.
AI Text Humanizer
Transform AI-generated text into natural, engaging writing with our Free AI Text Humanizer. Paste your text and see the magic happen in seconds!
Conclusion Generator
Craft compelling conclusions that leave a lasting impression with our user-friendly tool. Elevate your writing and make a lasting impact!
Lorem Ipsum Generator
Need a placeholder text for your design project? Our Lorem Ipsum Generator has got you covered! Create realistic and engaging placeholder text in seconds.
Paragraph Generator
Kickstart your writing process with our Paragraph Generator! Create fully coherent and compelling paragraphs that engage your readers.
Paraphrasing Tool
Quickly rephrase and reword any text with our Paraphrasing Tool. Perfect for essays, articles, emails, and more!
Sentence Rewriter Tool
Enhance the quality and clarity of any sentence with our Sentence Rewriter Tool. Improve your writing and make a lasting impact!
Grammar Checker
Catch grammar, spelling, and punctuation errors effortlessly and ensure your content is polished and error-free.
Emoji Translator
Add emotions to your text with our AI-powered Emoji Translator! Convert text to expressive emojis and communicate with feelings like never before.
Outline Generator
Turn your ideas into a clear and organized outline with our intuitive Outline Generator. Get started with your writing project in minutes!
Paragraph Rewriter
Improve the readability and clarity of any paragraph with our powerful Paragraph Rewriter. Make your writing sound more human-like and engaging!
Rewording Tool
Reword and rephrase sentences or paragraphs with ease using our Rewording Tool. Ideal for posts, emails, articles, and more!
Summarizer Tool
Experience the power of efficiency with our free Summarizer Tool! Simplify content, save time, and read smarter. Get instant results and elevate your writing!
Get Instant Access to 50+ Free AI Writing Tools
Take your writing to the next level with our extensive library of free AI writing tools.

Testimonials
Discover why academics trust Blainy to elevate their literature reviews.
Powered by Blainy . Elevate your writing skills with the industry’s favorite writing tool.
Elevate Your Research Today
Experience the future of literature review writing. Save time, gain deeper insights, and produce high-quality reviews with Blainy.

Unlock effortless writing excellence with the world's #1 AI-powered essay and research paper writer. Experience instant research paper perfection and elevate your writing to the next level.
Discover more.
50+ Free AI Tools
Terms & Condition
Privacy Policy
✉ [email protected]
✆ +971 50 760 0820
📍190 Hackett Inlet, Eastern Region, Dubai, UAE.
Copyright © 2024 Blainy
An official website of the United States government
Official websites use .gov A .gov website belongs to an official government organization in the United States.
Secure .gov websites use HTTPS A lock ( Lock Locked padlock icon ) or https:// means you've safely connected to the .gov website. Share sensitive information only on official, secure websites.
- Publications
- Account settings
- Advanced Search
- Journal List
Application of Artificial Intelligence in Pediatric Dentistry: A Literature Review
Noura alessa.
- Author information
- Article notes
- Copyright and License information
Address for correspondence: Dr. Noura Alessa, Department of Pediatric Dentistry and Orthodontics, Dental College, King Saud University, Riyadh, Saudi Arabia. E-mail: [email protected]
Received 2024 Feb 2; Revised 2024 Feb 11; Accepted 2024 Feb 12; Issue date 2024 Jul.
This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Artificial intelligence (AI) is the development of computer systems that can do tasks that normally require human intelligence. A number of dental specialties, including pediatric dentistry, now use AI and its subsets, machine learning, and deep learning. The evolution of AI in healthcare has been linked to the creation of AI applications meant to support medical professionals in diagnosing patients and choosing the best course of treatment. AI is the capability of robots to learn and use that information to carry out a range of cognitive tasks, including language processing, learning, reasoning, and making decisions—basically imitating human behavior. This review gives an overview of the numerous applications of AI that are beneficial to pediatric dentistry.
K EYWORDS: Artificial intelligence , artificial neural network , deep learning , pediatric dentistry
I NTRODUCTION
The early detection, management, and prevention of these problems are essential for a child’s best oral health. Artificial intelligence (AI) has demonstrated its use in the dental and medical fields in recent years.[ 1 ] The process of teaching a machine to think like a person is known as artificial intelligence or AI.[ 1 , 2 ] John McCarthy first used the term “artificial intelligence” in a 1956 conference at Dartmouth.[ 1 , 3 , 4 ] AI tools are becoming more and more important in numerous dental specialties these days. The development of AI programs meant to assist clinicians in diagnosing patients, selecting treatments, and forecasting outcomes has been linked to the application of AI in healthcare.[ 2 ]
The area of AI known as machine learning (ML) uses algorithms to forecast results based on a collection of data.[ 2 ] By utilizing data to construct algorithms, machines are able to tackle prediction issues without the assistance of humans. Artificial neurons are used by a collection of algorithms called neural networks (NNs) to compute signals. Artificial neurons that resemble human NNs are used in NNs, which use mathematical models to simulate the human brain. NNs have the ability to replicate human cognitive functions, such as problem-solving and thinking, learning, and making decisions, to mention a few. In essence, NNs consist of three layers: the input layer, which receives input from the user, the hidden layer, which processes data, and the output layer, which is where the system makes decisions.[ 5 ] Artificial neural networks (ANN), convolutional neural networks (CNN), and recurrent NNs are the three most used types of NNs. NNs include deep learning, which allows the computer to learn how to analyze data on its own. The hidden layer of deep learning NNs can have anything from a few thousand to a few million neurons.[ 2 , 6 , 7 ] NN helps to train computers to respond appropriately to events, instead of dictating what needs to be done.[ 8 ] More advanced AI solutions are more suited for use with 3D CNN for clinical reasons in dentistry (e.g., cone beam computed tomography or CBCT). CBCT, which requires significant radiation dosages, can be replaced in the field of endodontics by three CNNs, which can also identify anatomical features and tooth cavities. They are also essential in the field of oral pathology.[ 1 ]
Artificial narrow intelligence (ANI), artificial general intelligence (AGI), and artificial super intelligence (ASI) are the three main categories into which AIs are typically divided. ANI, also referred to as weak AI, has limited skills appropriate for extremely certain tasks. These systems only carry out the one function for which they were intended. Known as “strong” or “deep AI,” AGI is able to solve issues just like a human. In dentistry, NNs can be utilized to improve diagnosis accuracy, speed, and efficiency.[ 3 ]
Classification of AI
AI can be achieved in a variety of ways; different kinds of AI can accomplish varying jobs, and academics have developed a variety of AI categorization techniques. All non-human intelligence is referred to as AI. There are two further categories for AI: weak AI and strong AI. Strong AI is defined as AI with capabilities and intelligence on par with humans. Strong AI seeks to develop a decision-making multitasking algorithm. Expert systems and ML are two distinct subsets of weak AI. Currently, deep learning—a subset of ML—is one of the most popular research areas. One sort of deep learning model that is primarily utilized for picture production and recognition is CNNs. One type of deep learning algorithm is generative adversarial networks, an unsupervised learning technique intended to automatically identify patterns in the input data and produce new data with comparable properties.[ 9 ]
Evaluation of the present uses of AI in pediatric dentistry is lacking. Hence, the purpose of this review is to update about how useful AI as a diagnostic tool in pediatric dentistry.
AI advantages
ML and deep learning are two subgroups of AI, which have shown to be dependable additions to a clinician’s in decision-making process.
They provide improved monitoring, efficiency, accuracy, and precision, as well as time savings.
Reduces duration of investigation.
Feasible to improve people’s health at lower costs.
Offers personalized, predictive and preventative dentistry.
Disadvantages
There are few datasets and AI-based models available that can identify images that two-dimensional panoramic radiography cannot reach.
A single institution’s datasets are not widely available.
AI application in pediatric dentistry
AI offers a range of applications that span diagnosis, decision-making, treatment planning, and outcome prediction. Diagnosis, in particular, has seen significant improvements through AI.[ 10 ]
Detection of dental caries
AI algorithms can segment teeth, identify caries, and offer valuable predictions. AI can assist in diagnosing common pediatric dental issues, such as cavities, by analyzing X-rays and intraoral images, in educating young patients.[ 10 ] Talpur et al . conducted a study wherein they utilized deep-learning techniques to diagnose dental cavities through image analysis.[ 11 ] The edges of anatomical and pathological structures can be found using algorithms. To ascertain whether or not there were caries, an ANN was employed.[ 3 ]
With the use of straightforward questionnaires and tests, ML-based models assist in anticipating the existence of early childhood caries (ECC) in preschool-aged children. A new caries risk prediction model that considered genetic and environmental factors was developed. The most important items from the parent questionnaire are selected using an ML technique called random forest throughout the COVID-19 period in order to forecast the existence of active caries.[ 1 ] Karhade et al . used a questionnaire and ML to diagnose dental caries. The findings showed that ECC may be identified and classified with good accuracy.[ 12 ]
In assessing the child’s oral health and management
AI Identifies potential risk factors in pediatric oral health. It revolutionizes how data are collected, organized, and utilized enhancing the quality of care provided to children and adolescents. AI’s efficient data management capabilities provide pediatric dentists with a structured and centralized system for organizing the extensive medical records of children. This ensures quick access to crucial information about a child’s dental history, allowing for more personalized and child-centric care plans.[ 10 ] Wang, Y. et al . have pioneered the development of a comprehensive toolkit utilizing AI for the assessment of a child’s oral health.[ 4 , 10 ] ML models and algorithms improve the understanding and cognitive capabilities of dental professionals. They analyze patient data, medical records, and other relevant information to make predictions and treatment recommendations.[ 10 ] AI has applications in orthodontics for treatment planning, locating multiple cephalometric landmarks, and predicting treatment results.[ 9 ]
Fissure sealant categorization[ 1 ]
Adjusting dental sealants are identified using CNN. A high diagnosis accuracy was obtained by an AI-based system in contrast to the standard CNN-based classifications.
Age assessment in kids
In order to establish the chronological age of children and adolescents between the ages of 4 and 15, Zaborowicz, M. et al . employed three deep NN models. Their findings demonstrated that neural modeling algorithms could accurately determine metric age utilizing proprietary teeth and bone indicators.[ 13 ]
Identification of tooth and anomalies
Mesioden diagnosis using a single deep learning model. A deep learning method is used to identify and help with the early detection of germs in permanent or deciduous teeth.[ 1 , 14 ] One of the most widely used deep learning architectures, CNN, is frequently employed for object recognition. Pediatric kids’ deciduous teeth are increasingly being evaluated and counted using deep learning techniques like CNN. R-CNN inception has demonstrated good tooth identification accuracy. The first permanent molar ectopic eruption has been accurately identified by AI.[ 1 ] According to Bulatova et al. , the AI-based model was more efficient at identifying cephalometric landmarks than manual tracing.[ 15 ]
Endodontics
AI can be helpful in evaluating the anatomy of the root canal system, identifying working length measures, diagnosing periapical diseases and root fractures, and forecasting the outcome of retreatment treatments.[ 3 ] When testing three different CNNs for the diagnosis of pulpitis and deep caries on intraoral periapicals, Zheng et al . discovered that the multimodal CNN performed well in terms of accuracy.[ 16 ]
General dentistry
AI can help with precise shade matching.[ 3 ] It has been shown that CNN algorithms are useful tools for automatically identifying cancer and periodontal disease.[ 9 ] According to what you and your colleagues found, CNNs were able to identify dental plaque on primary teeth using intraoral photos with clinically meaningful findings.[ 17 ] Painless procedure: local anaesthesia can be achieved with AI-supervised nanorobotic anesthesia.[ 4 ]
The primary obstacles to applying AI in dentistry include using AI algorithms in the healthcare sector, which presents considerable hurdles, particularly with regard to the interchange and storage of clinical data. While AI cannot completely replace a dentist’s diagnostic procedure, it can help a general or pediatric dentist diagnose patients more quickly and with less error.[ 1 ]
C ONCLUSION
Without question, AI is a useful and potent tool for helping pediatric dentists. As a diagnostic tool, it has good levels of sensitivity, specificity, and accuracy. Still, more investigation is needed.
Financial support and sponsorship
Conflicts of interest.
There are no conflicts of interest.
Acknowledgment
Author would like to thank the college of dentistry Research centre and Deanship of Scientific Research at King Saud University for funding this project.
R EFERENCES
- 1. Vishwanathaiah S, Fageeh HN, Khanagar SB, Maganur PC. Artificial intelligence its uses and application in pediatric dentistry:A review. Biomedicines. 2023;11:788. doi: 10.3390/biomedicines11030788. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 2. Mahajan K, Kunte SS, Patil KV, Shah PP, Shah RV, Jajoo SS. Artificial intelligence in pediatric dentistry –A systematic review. J Dent Res Rev. 2023;10:7–12. [ Google Scholar ]
- 3. Lamba J, Malhotra T, Palwankar D, Vats V, Sachdeva A. Artificial intelligence in dentistry:A literature review. Biomed J Sci and Tech Res. 2023;51 [ Google Scholar ]
- 4. Magdline A, Moses J, Ranj E. Artificial Intelligence in Pediatric Dentistry:A Review. International Journal of Research Publication and Reviews. 2023;4(6):4256–4258. [ Google Scholar ]
- 5. Schwendicke F, Samek W, Krois J. Artificial Intelligence in Dentistry:Chances and Challenges. J Dent Res. 2020;99:769–74. doi: 10.1177/0022034520915714. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 6. Thurzo A, Urbanová W, Novák B, Czako L, Siebert T, Stano P, et al. Where is the artificial intelligence applied in dentistry?systematic review and literature analysis. Healthcare (Basel) 2022;10:1269. doi: 10.3390/healthcare10071269. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 7. Primer A Artificial Intelligence Versus Neural Networks. The Scientist Magazine®. [[Last accessed on 01 Feb 2024]]. Available from: https://www.the-scientist.com/magazine-issue/artificial-intelligence-versus-neural-networks-65802 .
- 8. Ghods K, Azizi A, Jafari A, Ghods K. Application of Artificial Intelligence in Clinical Dentistry, a Comprehensive Review of Literature. J Dent (Shiraz) 2023;24:356–71. doi: 10.30476/dentjods.2023.96835.1969. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 9. Ding H, Wu J, Zhao W, Matinlinna JP, Burrow MF, Tsoi KH. Artificial intelligence in dentistry—A review. Frontiers in Dental Medicine. 2023:1–13. Feb. [ Google Scholar ]
- 10. Deepika Lakshmi R, Brindha S, Dakshitha S, Deepak N, Moses J, Ranj E. Artificial Intelligence in Pediatric Dentistry:A Review. International Journal for Multidisciplinary Research (IJFMR) 2023;5(6.1) [ Google Scholar ]
- 11. Talpur S, Azim F, Rashid M, Syed SA, Talpur BA, Khan SJ. Uses of different machine learning algorithms for diagnosis of dental caries. J Healthc Eng. 2022;2022:5032435. doi: 10.1155/2022/5032435. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 12. Karhade DS, Roach J, Shrestha P, Simancas-Pallares MA, Ginnis J, Burk ZJS, et al. An automated machine learning classifier for early childhood caries. Pediatr Dent. 2021;43:191–7. [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 13. Zaborowicz M, Zaborowicz K, Biedziak B, Garbowski T. Deep learning neural modelling as a precise method in the assessment of the chronological age of children and adolescents using tooth and bone parameters. Sensors (Basel) 2022;22:637. doi: 10.3390/s22020637. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 14. Kılıc MC, Bayrakdar IS, Çelik Ö, Bilgir E, Orhan K, Aydın OB, et al. Artificial intelligence system for automatic deciduous tooth detection and numbering in panoramic radiographs. Dentomaxillofac Radiol. 2021;50:20200172. doi: 10.1259/dmfr.20200172. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 15. Bulatova G, Kusnoto B, Grace V, Tsay TP, Avenetti DM, Sanchez FJC. Assessment of automatic cephalometric landmark identification using artificial intelligence. Orthod Craniofac Res. 2021;24(Suppl 2):37–42. doi: 10.1111/ocr.12542. [ DOI ] [ PubMed ] [ Google Scholar ]
- 16. Zheng L, Wang H, Mei L, Chen Q, Zhang Y, Zhang H. Artificial intelligence in digital cariology:A new tool for the diagnosis of deep caries and pulpitis using convolutional neural networks. Ann Transl Med. 2021;9:763. doi: 10.21037/atm-21-119. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- 17. You W, Hao A, Li S, Wang Y, Xia B. Deep learning-based dental plaque detection on primary teeth:A comparison with clinical assessments. BMC Oral Health. 2020;20:141. doi: 10.1186/s12903-020-01114-6. [ DOI ] [ PMC free article ] [ PubMed ] [ Google Scholar ]
- View on publisher site
- Collections
Similar articles
Cited by other articles, links to ncbi databases.
- Download .nbib .nbib
- Format: AMA APA MLA NLM
Add to Collections
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 03 November 2024
Multimodal AI/ML for discovering novel biomarkers and predicting disease using multi-omics profiles of patients with cardiovascular diseases
- William DeGroat 1 na1 ,
- Habiba Abdelhalim 1 na1 ,
- Elizabeth Peker 1 na1 ,
- Neev Sheth 1 na1 ,
- Rishabh Narayanan 1 na1 ,
- Saman Zeeshan 2 ,
- Bruce T. Liang 3 , 4 &
- Zeeshan Ahmed 1 , 4 , 5 , 6
Scientific Reports volume 14 , Article number: 26503 ( 2024 ) Cite this article
Metrics details
- Cardiovascular diseases
- Personalized medicine
Cardiovascular diseases (CVDs) are complex, multifactorial conditions that require personalized assessment and treatment. Advancements in multi-omics technologies, namely RNA sequencing and whole-genome sequencing, have provided translational researchers with a comprehensive view of the human genome. The efficient synthesis and analysis of this data through integrated approach that characterizes genetic variants alongside expression patterns linked to emerging phenotypes, can reveal novel biomarkers and enable the segmentation of patient populations based on personalized risk factors. In this study, we present a cutting-edge methodology rooted in the integration of traditional bioinformatics, classical statistics, and multimodal machine learning techniques. Our approach has the potential to uncover the intricate mechanisms underlying CVD, enabling patient-specific risk and response profiling. We sourced transcriptomic expression data and single nucleotide polymorphisms (SNPs) from both CVD patients and healthy controls. By integrating these multi-omics datasets with clinical demographic information, we generated patient-specific profiles. Utilizing a robust feature selection approach, we identified a signature of 27 transcriptomic features and SNPs that are effective predictors of CVD. Differential expression analysis, combined with minimum redundancy maximum relevance feature selection, highlighted biomarkers that explain the disease phenotype. This approach prioritizes both biological relevance and efficiency in machine learning. We employed Combination Annotation Dependent Depletion scores and allele frequencies to identify variants with pathogenic characteristics in CVD patients. Classification models trained on this signature demonstrated high-accuracy predictions for CVD. The best performing of these models was an XGBoost classifier optimized via Bayesian hyperparameter tuning, which was able to correctly classify all patients in our test dataset. Using SHapley Additive exPlanations, we created risk assessments for patients, offering further contextualization of these predictions in a clinical setting. Across the cohort, RPL36AP37 and HBA1 were scored as the most important biomarkers for predicting CVDs. A comprehensive literature review revealed that a substantial portion of the diagnostic biomarkers identified have previously been associated with CVD. The framework we propose in this study is unbiased and generalizable to other diseases and disorders.
Introduction
Cardiovascular diseases (CVDs) are recognized as the primary cause of mortality among men and women in the United States 1 , 2 . Given their complex nature, risk factors, inherent genetic makeup, and trajectory, personalized management is essential for the effective treatment of CVDs 2 . Advancements in genomics and bioinformatics have significantly enhanced our understanding of the intricate origins of CVDs 3 , 4 . Gaining insights into disease implications by utilizing transcriptomic expression and variant profiles holds the promise of revolutionizing diagnostic capabilities, treatment strategies, and prognostic assessments across various CVDs including but not limited to heart failure (HF) and atrial fibrillation (AF) 5 , 6 . These advancements stem from next-generation sequencing (NGS) technologies, which have facilitated the identification of novel heritable links and the exploration of genetic diversity among patients 7 . Gene expression analysis through RNA-sequencing (RNA-seq) data has aided in uncovering disease associated biomarkers and categorizing patient groups according to their risk profiles 8 . Analyzing RNA-seq data for differential expression allows for the exploration of genome-wide biological disparities, leading to enriched functional pathways and gene ontologies 9 , 10 .
RNA-seq datasets provide valuable biological insights into gene expression, RNA processing, and molecular pathways underlying disease states 11 , 12 . While gene expression analysis allows for enhancements in diagnostic capabilities and precise treatment plans, multiple studies have established that RNA-seq provides limited coverage of non-coding regions and that transcriptomics cannot detect genomic variants 11 , 12 , 13 . The onset of multifactorial diseases is shaped by an interplay of environmental and genetic factors, affecting various biological processes such as gene regulation 14 . Previous studies utilizing whole genome and exome sequencing (WGS/WES) have demonstrated their effectiveness in accurately revealing the effects of non-coding variants on CVDs 15 , 16 and other complex diseases 17 , as well as in capturing all genetic variation, thus, providing comprehensive information about an individual’s entire genome 16 , 17 . Although sequencing technology aids in identifying genetic variations linked to diseases, accurately linking specific genomic variations to disease phenotypes remains challenging 3 , 18 . Deciphering the pathogenic and biological function of genes may require additional information beyond what a single type of data can offer 18 . Data integration is vital in managing the escalating volume of data and obtaining comprehensive interdisciplinary insights into extensive genomic datasets 19 . Additionally, due to the heterogenous nature of genomic, transcriptomic, and clinical data, a lack of standardization persists as a limitation in data integration 18 . These challenges are being addressed with the integration of precision medicine and artificial intelligence (AI)/machine learning (ML) approaches where phenotypic, clinical, transcriptomic, and genomic data can be selected and classified to facilitate the identification of high-risk patients 18 , 20 . Utilizing cutting edge AI/ML technology can aid in the analysis and interpretation of gene expression and variant data, providing more accurate diagnosis and improving our understanding of the mechanisms behind complex diseases including but not limited to CVDs 21 , 22 , lupus 23 , and colon cancer 24 .
Previously, we conducted traditional bioinformatic analyses, including an in-depth gene expression and enrichment analysis of RNA-seq data from patients with primarily HF and other CVDs. We identified differentially expressed genes (DEGs) that are well-documented to be associated with CVDs and other enriched pathways 25 . However, we were unable to detect any CVD drivers using RNA-seq data. To address this limitation, we employed an integrative, multi-omics approach incorporating gene expression, pathogenic genetic variants, and associated phenotypes among CVD populations 6 . In this study, we combined specific mutations for the DEGs we had previously reported, allowing for a better understanding of CVD progression 6 . Extending our research and expanding beyond orthodox bioinformatic techniques, we implemented AI/ML techniques on RNA-seq-driven gene expression data to study biomarkers associated with HF, AF, and other CVDs 26 . Our AI/ML analysis supported our initial gene expression study as we were able to identify common genes that have a high impact on CVD diagnosis 25 , 26 . Additionally, this AI/ML framework aided in establishing Hygieia , a portable pipeline that integrates genomics and healthcare data to explore genes linked to specific disorders and predict disease 27 . While we were able to predict CVDs with high accuracy using this methodology, we were only focused on CVD driver genes with genetic alterations that can culminate in CVD 26 . We overcame this challenge by using whole-transcriptome-based gene expression data and further enhanced our AI/ML model to encompass a novel nexus of algorithms to predict CVDs based on crucial transcriptomic biomarkers 28 . We utilized this approach and proposed IntelliGenes , a novel AI/ML pipeline for the identification of novel biomarkers and the training of single-disease prediction models 29 .
In this study, we leverage our previous work and present a new AI/ML approach that uses multi-omics data, integrating RNA-seq-driven gene expression, whole genome-based single nucleotide polymorphisms (SNPs), and clinical demographics data (Fig. 1 ). Biomarkers associated with CVDs based on differential expression were investigated for pathogenic SNPs within the genes and their regulatory elements. A clinically integrated genomic and transcriptomic (CIGT) dataset was analyzed using three ML algorithms to accurately predict CVDs. Through the identification of genetic biomarkers and their associated SNPs, we have highlighted potential indicators for the early detection of CVDs. These biomarkers aid in identifying individuals at risk before diagnosis, enabling prompt intervention and enhancing patient outcomes. With its implementation in healthcare, our predictive model can identify patients at risk for CVDs and may be adapted to perform other single disease predictions.
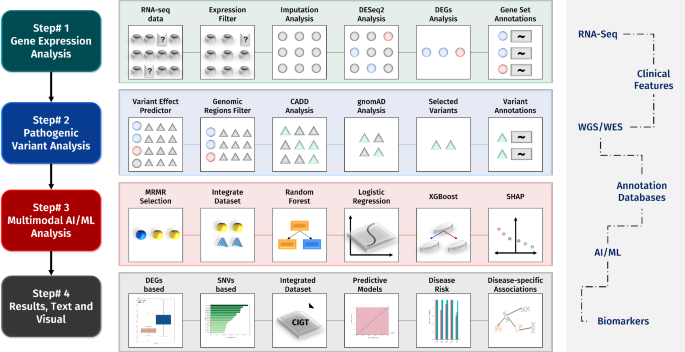
Study design and workflow. This figure represents a summary of our study design: (I) Transcriptomic expression, (II) Pathogenic Variant, (III) Multimodal Machine Learning, and (IV) Results. Various inputs and their implementation are also included (RNA-sequencing, Clinical Records, Whole Genome Sequencing, Annotation databases and Biomarkers).
Our methodology is separated into four steps (Fig. 2 ): (I) Pre-processing, (II) Transcriptomic/gene expression analysis, (III) Pathogenic variant analysis, and (IV) Multimodal AI/ML analysis.
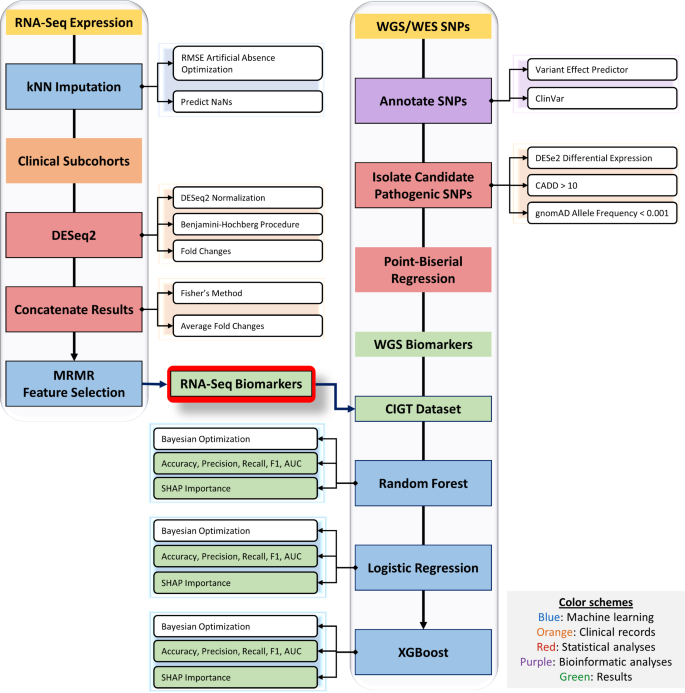
Methodology. This figure presents the k-Nearest Neighbors (k-NN) imputation to address missing values present in our RNA-seq expression data. DESeq2 was utilized for normalization and gene differential expression on four clinical sub cohorts to reduce the effect of confounding variables. Next, minimum redundancy – maximum relevance (MRMR) was performed to identify biomarkers proficient in predicting CVDs. Simultaneously, significant single nucleotide variants (SNVs) were annotated, and their pathogenicity determined for downstream analysis. Utilizing the clinically integrated transcriptomics and genomics dataset (CIGT) of significant biomarkers and their variants, machine learning algorithms (Random Forest, Logistic Regression, and Xtreme Gradient Boosting) to predict CVDs. Boxes highlighted in yellow refer to input data, blue refers to machine learning approaches, orange highlights clinical records, red refers to statistical analyses, while purple refers to bioinformatic analyses, and green highlights results.
Pre-processing
Previously, we performed RNA-seq on a 71-participant cohort of CVD patients and healthy controls 6 , 25 . Samples were collected from the individuals’ peripheral blood mononuclear cells (PBMCs). All procedures performed in studies involving human participants were in accordance with the ethical standards of the institution and the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All human samples were used in accordance with relevant guidelines and regulations, and all experimental protocols were approved by the Institutional Review Board at UConn Health and Rutgers. This cohort included 61 CVD patients: 40 male and 21 female individuals aged between 45 and 92, from diverse ethnic groups (56 non-Hispanic, four Hispanic, and one declined to answer) and self-described race (42 White, seven Black or African American, one Asian and 11 of unknown race). Ten control participants rounded out the cohort: five males and five females (of whom three were self-described Hispanics and seven non-Hispanic; nine were White race and one unknown race) aged between 28 and 78 with no clinical manifestation of CVD. From our 71-participant cohort, we extracted transcriptomic expression for each individual. Counts and transcripts per million (TPM) values were retrieved from our RNA-seq data. TPM values guided preprocessing. Transcripts with a median TPM below 0.5 across participants were removed from our dataset, and transcripts without significant expression (TPM > 1) in at least one patient were also removed. Additionally, non-ubiquitous transcripts, or transcripts expressed in less than 80% of the cohort participants’ RNA-seq, were excluded from downstream analysis. The raw counts of the filtered transcripts were configured into a CIGT-formatted dataset based on raw count values; this dataset served as the basis for our subsequent analyses.
We applied k-nearest neighbors (k-NN) imputation paired with artificially induced missingness to predict missing count values within our CIGT-formatted dataset (e.g., NaN). Here, we replaced 10% of the known portion of our dataset with missing values, maintaining the true values for comparison. We simulated imputations using distinct ‘n_neighbors’ values, ranging from 1 to 20. Optimizing this parameter assists in reducing noise in high-dimensional datasets, fine-tuning the imputer’s sensitivity; essentially, this parameter controls the number of most similar data points to consult when estimating a missing value. With each simulation, we calculated the root mean squared error (RMSE) by comparing the predicted values with the previously withheld portion of the dataset; RMSE was minimized to determine our choice of optimal ‘n_neighbors’ values. We filled in missing values using optimal ‘n_neighbors’ and reintegrated the artificially excluded values into our dataset. Imputation is necessary, as DESeq2 requires the input RNA-seq counts to be complete (i.e., not contain NaN values). We conducted dataset normalization using DESeq2’s median-of-ratios on our continuous imputed dataset 30 , 31 . Median-of-ratios normalization improves cross-sample comparability by using a robust, sample-wide scaling factor that reduces biases from sequencing depth and compositional differences, thereby maintaining biological signal integrity better than TPM normalization 32 . We calculated the coefficient of variation and intraclass correlation for our normalized dataset. Because median of ratios is a dataset-dependent normalization algorithm, subsets matrices derived from our expression data must be renormalized from raw counts prior to further analysis.
Transcriptomic/gene expression analysis
Next, we performed differential expression analysis using DESeq2. To minimize the effects of confounders, the cohort’s CVD patients and control participants were stratified into subcohorts based on their demographic features. We assigned groups using the individuals’ sex, racial background, and age. Four groups were created: white males aged 45–64, white males aged 65 and older, white females aged 45–64, and white females aged 65 and older (further details are available in Supplementary Material 1, attached). Limitations in our cohort’s construction prevented us from creating groupings for other races, as we lacked the necessary participants. Differential expression using DESeq2 was then performed independently on each subcohort, employing a negative binomial distribution-based model to identify DEGs 30 . Each analysis on the subcohorts yielded separate adjusted p-values and log fold changes (LFC), respectively indicating the probability of each feature being differentially expressed and its expression direction. A positive LFC value indicates upregulation (i.e., overexpression), and a negative LFC indicates downregulation (i.e., underexpression). The results from our four subcohorts were merged: adjusted p-values were combined with Fisher’s method, and LFCs were averaged. Only DEGs with a newly calculated p-value less than 0.05 were considered significant and included in downstream analysis.
To determine biologically relevant DEGs for use in ML single-disease predictions 33 , 34 and to select genes that explain a generalized CVD phenotype, we utilized minimum redundancy - maximum relevance (MRMR). In MRMR, an arbitrary ‘k’ parameter is chosen, indicating the best set of ‘k’ size for explaining the difference between patients and controls. MRMR minimizes the effects of co-expressed DEGs and other patterns of redundant information contained in expression datasets. This approach is preferable to other ML selectors with arbitrary cutoffs, such as recursive feature elimination models. In these models, highly correlated biomarkers might aggregate at the top of rankings, leaving classifiers with less useful or more redundant information to make predictions. For our analysis, we performed MRMR exclusively on the DEGs in our training dataset, with ‘k’ set as 10. Using Gene Set Enrichment Analysis (GSEA), we examined Gene Ontology (GO) and Human Phenotype Ontology (HPO) enrichment for our 10 MRMR-selected biomarkers 35 , 36 , 37 . GO was utilized to investigate the biological processes, cellular components, and molecular functions of our biomarkers. HPO assisted in searching for disease implications.
Pathogenic variant analysis
WGS data from our 71-participant cohort was investigated alongside RNA-seq. We processed WGS-derived SNPs from each patient into VCF format. SNPs with quality scores below 50 in the VCF files were discarded. Using the Ensembl Variant Effect Predictor (VEP), we annotated these genomes with ClinVar, Combined Annotation Dependent Depletion (CADD), and Genome Aggregation Database (gnomAD). Here, we exclusively examined the WGS datasets of CVD patients in our training cohort for these SNPs. Only SNPs associated with our MRMR-selected DEGs and their known regulatory elements (i.e., promoters and enhancers), sourced from GeneHancer 38 , were included in downstream analyses. This methodology allowed us to focus on genomic regions we had previously been able to implicate with CVD. By excluding SNPs outside these key areas, our study minimizes confounding data, thereby improving the likelihood of identifying significant genetic contributors to CVD.
Next, we utilized CADD to measure the deleteriousness of SNPs in our regions of interest. CADD is a computational tool designed to evaluate the pathogenicity of SNPs 39 . CADD utilizes machine learning to integrate multiple genomic annotations, predicting the harmfulness of genetic variants. This tool leverages data from diverse sources, including evolutionary conservation and functional annotations, to generate a comprehensive score that assesses variant impact. As our threshold, we used a CADD PHRED score greater than 10, representing the top 10% of harmful SNPs, as our threshold 39 . To identify rare variants within this highly deleterious set, we used gnomAD, a comprehensive public resource that aggregates exome and genome sequencing data to provide insights into genetic variants across diverse populations. It offers critical information on the frequency and potential impact of genetic mutations. Specifically, gnomAD provided allele frequencies for SNPs; variants with an allele frequency of < 0.1% were included in our analyses 40 . We then assessed the presence of SNPs from our training dataset in the rest of our 71-patient cohort. Their presence/absence was detailed in a binary matrix in the CIGT format. This matrix was merged with our RNA-seq counts matrix by a common ID identifying each patient.
Multimodal AI/ML analysis
Our selected DEGs and SNPs were integrated into an AI/ML-ready CIGT-formatted dataset and used with an 80/20 train-test split by three ML classifiers to predict CVD risk. Using Bayesian optimization, we found optimal hyperparameters for random forest (RF), eXtreme Gradient Boosting (XGBoost), and logistic regression (LR) models. For RF, ‘max_depth,’ ‘min_samples_split,’ ‘min_samples_leaf,’ ‘n_estimators,’ ‘max_features,’ ‘bootstrap,’ ‘criterion,’ ‘max_leaf_nodes,’ and ‘max_samples’ were optimized. Our XGBoost classifier was optimized for ‘max_depth,’ ‘min_child_weight,’ ‘gamma,’ ‘subsample,’ ‘colsample_bytree,’ ‘scale_pos_weight,’ ‘n_estimators,’ and ‘learning_rate.’ The parameters ‘C,’ ‘penalty,’ ‘solver,’ and ‘max_iter’ were included in the Bayesian optimization for LR. The Bayesian optimization was performed with 10-fold cross-validation. Optimizing these hyperparameters assists in maximizing the performance of each classifier on the dataset. Bayesian optimization is a specific sequential optimization technique that enables faster convergence of parameters over typical brute force algorithms such as Grid Search. RF and XGBoost, two tree-based models, were chosen as they have previously proven powerful in single-disease prediction 28 . LR, a linear model, was chosen for comparison. These classifiers performed patient-specific single-disease prediction on our testing dataset. Metrics detailing the classifiers’ performance were computed: accuracy, AUC, probabilities, sensitivity, specificity, F1, and Brier score. To investigate the importance and directionality of each feature in predicting the CVD phenotype for each ML model, SHapley Additive exPlanations (SHAP) were computed for each feature. SHAP scores offer insights into patient-specific CVD manifestations using a game-theoretic approach 41 . We combined the SHAP profiles with prediction probabilities to investigate which biomarkers were the most important contributors to each patient’s CVD prediction. Additionally, we conducted an extensive literature review to examine which of these biomarkers have previously been implicated in manifestations of CVD.
We designed comparable, RNA-seq data-driven, patient-specific expression profiles for efficient DE and ML analyses 29 . Datasets are available in Supplementary Material 2. We identified 56,681 initial transcripts in our cohort. Pre-processing revealed 748 transcripts suitable for downstream differential expression analysis (DEA). Missing expression values in individual patients’ profiles necessitated imputation before DEA. Using a robust, empirical approach to parameterize the imputer, we addressed dataset gaps. Artificial missingness facilitated RMSE calculations across various simulations of ‘n_neighbors.’ k-NN Imputation was performed using our optimal ‘n_neighbors’ of 11, which had the lowest RMSE across simulations. With a completed dataset, DESeq2 normalization was performed for improved cross-sample comparability. Through a demographic-based segmentation approach, we performed DEA to investigate potential CVD-associated transcripts across four subcohorts, minimizing noise from confounding variables. The subcohort-specific results were then merged, detecting 28 DEGs, detailed in Table 1 , across our cohort. LFCs characterized each DEG’s direction of regulation in CVD patients. Here, we demonstrate 20 upregulated DEGs and 8 downregulated DEGs.
We implemented MRMR feature selection to capture the phenotypic profile of CVD in 10 DEGs: ITGB2 , CD37 , RPL36AP37 , PSAP , ACTB , SELL , NCF2 , HBA1 , ICAM3 , and BBLN . This approach excludes transcriptomic features containing non-informative and redundant information for ML classifiers (e.g., co-expression). As demonstrated, these 10 DEGs successfully explain the differences between CVD patients and healthy controls. Figure 3 A and B detail each DEG’s LFC and adjusted p-value. Additionally, we examined GO and HPO enrichment across the transcript set to examine their implicated pathways and clinical relevance (Fig. 3 C). Using Fisher’s exact test, we conclude that downregulated DEGs are enriched in our MRMR-selected DEGs, with a p-value of 0.022. In authentic literature, seven of the MRMR-selected transcripts were associated with CVDs. Loss of HBA1 function is linked with coronary artery disease (CAD) 42 . Hypomethylation of ITGB2 in PBMCs is linked to HF and CAD 43 , 44 . SNPs affecting SELL are associated with an increased risk of acute coronary syndromes (ACSs) 45 . Blood-based hypermethylation of ACTB is associated with the development of coronary heart disease (CHD) 46 . Conversely, ACTB hypomethylation increases the risk of stroke 47 . NCF2 has been used as a diagnostic biomarker for obstructive CAD in PBMCs 48 . Additionally, NCF2 has been associated with AF 49 . ICAM3 has been identified as a prognostic biomarker for acute ischemic stroke 50 . BBLN has been found to be differentially expressed in damaged hearts 51 , 52 .
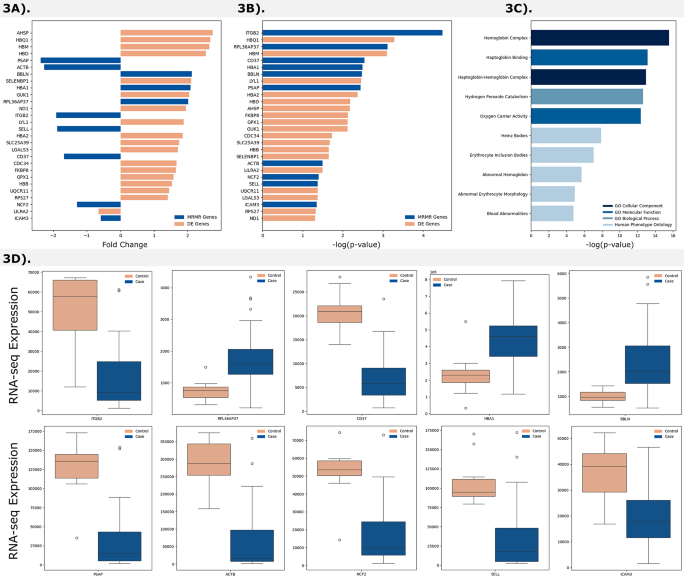
Differentially expressed genes and their expression plots. This figure presents the results of gene expression analysis and that includes, (A) Fold change in expression level based on differential expression (DE) analysis and redundancy – maximum relevance (MRMR) feature selection; (B) Significance levels of genes based on DE and MRMR; (C) Gene annotations for cellular component, molecular function, biological processes, and phenotypic abnormalities; and (D) RNA-seq expression plots for the ten most significant biomarkers.
We integrated rare, deleterious WGS-based SNPs into our patient-specific, ML-efficient profiling. We performed a rigorous search of DEGs and their regulators for SNPs with pathogenic characteristics. Our analysis revealed 17 SNPs matching these criteria in our cohort. Figure 4 A and B demonstrate the CADD PHRED scores and allele frequencies sourced from gnomAD for these SNPs. Additionally, in Table 2 , we report the location, transcripts, and consequences of the 17 SNPs. The distribution of consequences is shown in Fig. 4 C. The majority of the SNPs we located had not been previously reported or were scored with uncertain or conflicting pathogenicity in ClinVar. Only rs115891972 and rs751011909 were classified as benign and likely benign, respectively. Next, using an uncontaminated training dataset isolated from the testing dataset during feature selection and hyperparameter tuning, we trained three distinct ML classifiers (further details are available in Supplementary Material 3). Our features consisted of 10 MRMR-selected DEGs and 17 SNPs. The decision tree (DT) classifiers, RF and XGBoost, demonstrated perfect ability to differentiate between CVD patients and healthy controls, achieving 100% accuracy. Notably, we used a small testing set of 15 participants. Overall, XGBoost performed the best, considering the Brier score. Our LR model performed worse, scoring 93% accuracy but failing to detect the sole healthy individual in our testing dataset. Here, we conclude that DT models are more suitable for single-disease predictions. They provide interpretable ML models capable of handling non-linear relationships and synthesizing various variable types, which are strengths necessary for high-dimensional multi-omics datasets. Figure 5 A and B display our classifiers’ predictions and AUC-ROC. The integrated, multi-omics patient-specific profiles containing SNPs and gene expression data outperformed non-integrated RNA-seq datasets. Previously, we demonstrated 91% accuracy using a comparable RNA-seq dataset with RF and XGBoost classifiers trained on RNA-seq expression 28 .
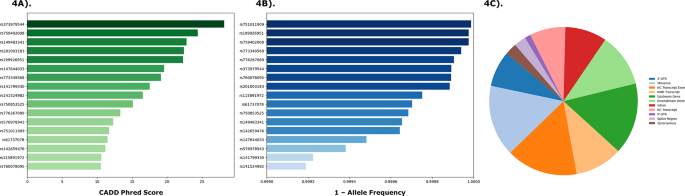
Variant feature selection. This figure presents the rare, deleterious variants affecting our CVD associated biomarkers based on, (A) Combined Annotation Dependent Depletion (CADD) Score; (B) Allele frequency obtained from the Genome Aggregation Database (gnomAD); and (C) annotations of pathogenic single nucleotide variants (SNVs).
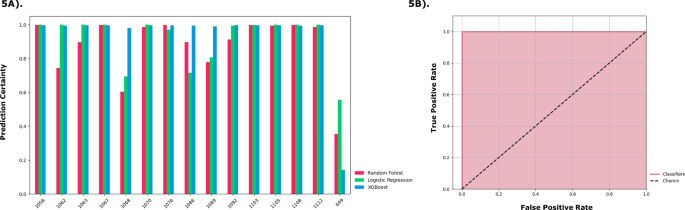
Predictive analysis. This figure presents the predictive confidence of our ML model and that includes, (A) Predictive certainty of three ML algorithms (Random Forest, Logistic Regression and Xtreme Gradient Boosting) on testing dataset; and (B) Receiver operating characteristic (ROC) curve denoting the sensitivity and specificity of the classifiers.
We created single-patient profiles of CVD phenotypes using SHAP importances and prediction metrics from our best-performing XGBoost model. Examination of these profiles revealed that RPL36AP37 (mean absolute SHAP value = 0.76) and HBA1 (mean absolute SHAP value = 0.52) demonstrated greater usefulness than other MRMR-selected biomarkers in training the classifier, with BBLN (mean absolute SHAP value = 0.22) as the next highest. ICAM3 was not utilized by our XGBoost algorithm. Biomarkers contributing more significantly to predictions could indicate disease involvement, leading to more efficient diagnoses and treatment of CVD.
This study explores the functional impact of ML-based multi-omics interactions in CVD. Our DEA identified 28 DEGs (Table 1 ), with 12 previously associated with a phenotypic variation of CVDs. Upon further investigation into CVD and non-CVD associations, we found that 65% of the total diseases reported for all genes were related to CVDs. Importantly, patients with comorbidities were not excluded when consenting to our cohort of CVD patients, a potential causal factor for non-CVD associations. Two genes ( HBM and GUK1 ) had exclusively CVD associations. The gene HBM has previously been identified as one of the ten most DEGs for hypertrophic cardiomyopathy (HCM) patients 53 . Additionally, it has been linked to CVD-related risk factors such as pulmonary arterial hypertension 54 and alpha-thalassemia 55 . Upregulation of GUK1 has been implicated in CHD and HCM 56 . Ten of the twenty-eight DEGs ( HBA1 , GPX1 , SELENBP1 , LGALS3 , ND1 , ITGB2 , ACTB , NCF2 , SELL , and ICAM3 ) were linked to both CVDs and non-CVDs. HBA1 has been documented to be highly associated with CVDs such as ischemic heart disease 57 and CAD 58 . It has also been linked to other CVD-related risk factors such as hypertension 59 and alpha-thalassemia 59 , as well as other non-CVDs such as chronic kidney disease, sickle cell disease 60 , and nonalcoholic fatty liver disease 61 . LGALS3 62 , ITGB2 44 , and ICAM3 50 were all found to be correlated with ischemic stroke and CHD. Upregulation of GPX1 has been implicated in various complex disorders, including but not limited to cardiomyopathy 63 , acute myeloid leukemia (AML) 63 , and endometrial cancer 64 . Additionally, upregulation of SELENBP1 has been associated with acute coronary syndrome 65 . It has also been implicated in other phenotypes of CVDs, such as myocardial infarction and cardiac arrest 65 , as well as non-CVDs, including breast cancer and lung adenocarcinoma 66 . Downregulation of ND1 67 and upregulation of ACTB 46 , 68 in the inflammatory pathways are known to be associated with CHD and cardiomyopathy. ND1 and ACTB are also linked to other chronic and heritable diseases, such as mitochondrial encephalomyopathy 69 and Parkinson’s disease 70 , respectively. Additionally, NCF2 and SELL are reported to be potential diagnostic biomarkers for CAD 48 , 72 , as well as cancers such as hepatocellular carcinoma 72 and leukemia 73 , respectively. While the direct correlation between other complex diseases and CVD remains unknown, state-of-the-art literature supports the implication of these genes in the inflammatory and immunological pathways shared between these diseases 59 , 60 , 66 , 73 . Future genomic and translational studies are required to understand these relationships.
Sixteen of the twenty-eight DEGs were found to be only associated only with non-CVDs based on existing literature (Table 1 ). Genes such as HBQ1 74 , HBA2 75 , CD37 76 , and LILRA2 77 are all associated with different types of cancer and other immunological diseases documented to directly impact CVD pathophysiology. While these genes are not directly linked to CVDs, further research is required to understand their effects on regulatory elements that might trigger CVD development. We could not find evidence linking genes such as BBLN , AHSP , and HBB with CVD or other diseases. However, we reported their implications in CVD risk factors, including but not limited to Tetralogy of Fallot 51 and beta thalassemia 78 , 79 , respectively. Other genes such as RPL36AP37 80 , LYL1 81 , HBD 82 , FKBP8 83 , CDC34 84 , SLC25A39 85 , UQCR11 86 , RPS27 87 , and PSAP 88 are all implicated in cancerous and neurological diseases that are not directly associated with CVDs. A detailed list of documented CVD and non-CVD phenotypes associated with our DEGs is available in Table 1 .
We validated our findings with peer-reviewed studies (Table 3 ). Eleven of the twenty-eight genes were found to be upregulated in our DEA as well as existing literature. HBM 53 , GUK1 56 , HBA1 58 , GPX1 63 , SELENBP1 65 , and LGALS3 62 were all upregulated in different phenotypes associated with CVDs. Other genes, such as HBQ1 74 , BBLN 51 , LYL1 81 , CDC34 84 , and UQCR11 86 , were implicated in other diseases, but their regulation levels also matched existing studies. Additionally, two genes, SELL 72 and CD37 76 , were observed to be downregulated in our DEA and in current studies. Seven of these genes ( HBA1 , RPL36AP37 , BBLN , ITGB2 , ACTB , NCF2 , SELL , ICAM3 , CD37 , and PSAP ) were selected using MRMR and later utilized by our AI/ML model to predict CVDs. Upregulation of HBA1 has been extensively reported to exhibit a strong correlation with ischemic heart disease 57 , while loss of HBA1 function is associated with CAD 55 . In our previous studies, we identified protein-coding HBA1 to be upregulated in CVD patients and significantly expressed in HF patients 14 , 25 , 26 . The pseudogene RPL36AP37 assists in regulating DNA replication within eukaryotic cells and in producing ribosomal proteins 89 . Little to no information linking RPL36AP37 to CVDs has been reported. However, non-CVD phenotypes associated with altered or loss of RPL36AP37 function include primary angle closure glaucoma 89 , Parkinson’s disease, and certain cancers 80 .
The protein-coding BBLN serves as a vital regulator of intestinal intermediate filaments, crucial for normal intestinal function, while also playing a role in maintaining cellular organelle architecture and serving as molecular spacers 90 . Induced downregulation of BBLN in mice with congenital heart defects leads to further cardiovascular dysfunction and necroptosis via activation of the CAMK2D pathway 51 . ITGB2 , a protein-coding gene, encodes a cell-associated signaling molecule involved in the leukocyte adhesion and migration of T-cells and neutrophils 91 . ITGB2 is highly expressed within AML patients 43 , and hypomethylation of this gene in peripheral blood was linked to HF and CAD 44 . Blood-based hypermethylation of ACTB , another protein-coding gene, was significantly associated with the development of CHD 46 . In contrast, hypomethylation of ACTB was found to increase the risk of stroke 92 . The protein-coding NCF2 was found to be significantly upregulated in AF 93 and significantly expressed in CAD patients 48 . SELL encodes for selectin, a protein essential for binding and rolling leukocytes on endothelial cells, and acts as a primary downstream target for DYSF , a protein that, if upregulated, contributes to the pathogenesis of atherosclerotic CVDs 94 . Upregulation of the protein-coding gene ICAM3 is identified as a prognostic biomarker for acute ischemic stroke 50 . CD37 , a protein-coding gene, regulates immune response and prevents tumor formation, with upregulation observed in mRNA expression levels is observed in AML patients 76 . PSAP , a protein-coding gene, plays a significant role in the pathogenesis of atherosclerosis, a key risk factor for CVDs 95 . Elevated PSAP expression in plaque macrophages was related to atherosclerosis-linked inflammation in humans 95 . Additionally, these seven genes are linked to other multi-factorial diseases. Further studies are needed to understand the non-CVD implications on CVD prognosis and diagnosis.
The integration of multi-omics data, coupled with advances in the multimodal advancements in AI/ML, has the potential to enhance diagnostic and predictive analyses of leading causes of mortality, modifiable risk factors, and other medical insights. State-of-the-art literature has supported the implementation of a genomic language model trained on millions of metagenomic scaffolds to uncover hidden functional and regulatory connections among genes. This process also reveals intricate relationships between genes within a genomic region 96 . Another recent study utilized a deep-learning, integrative mass spectrometry framework on metabolomics focused on lipid profiles to detect lipid content specific to regions and localize lipids to individual cells depending on both cell subpopulations and the anatomical origins of the cells 97 . In epigenetics, recent literature introduced DeepMod2, a comprehensive deep-learning framework designed for methylation detection using the ionic current signal obtained from Nanopore sequencing 98 . It incorporates both a bidirectional long short-term memory model and a transformer model to facilitate rapid and precise detection of DNA methylation from different flow cell types using WGS or adaptive sequencing data 98 . Another study showcased a multi-omics analytic platform leveraging genomic, transcriptomic, proteomic, and lipid data to accurately predict adenocarcinoma patient survival 99 . This platform employs an ensemble of algorithms, including support vector machine, random forest, and neural network, to identify disease-associated biomarker panels for downstream predictive analyses 99 . Furthermore, researchers have combined omics data with demographic and clinical information to offer a comprehensive view of cancer prognosis 100 . They created and validated a deep learning framework capable of extracting insights from complex gene and miRNA expression data, enabling accurate prognosis predictions for breast and ovarian cancer patients 100 . All of these approaches offer potential advancements in understanding disease biology and could assist in developing more targeted treatments.
Data availability
Processed data and related material are attached in the supplementary material. All the source code reproducing the experiments of this study are available at GitHub, following web link: < https://github.com/drzeeshanahmed/intelligenes_multi-omics_cvd_analysis>.
Abbreviations
Acute Coronary Syndrome
- Artificial Intelligence
Atrial Fibrillation
Acute Myeloid Leukemia
Area Under the Curve
Cardiovascular Disease
Coronary Artery Disease
Coronary Heart Disease
Combined Annotation Dependent Depletion
Clinically Integrated Genomic and Transcriptomic
Decision tree
Differentially Expressed Genes
Gene Ontology
Gene Set Enrichment Analysis
Genome Aggregation Database
Heart Failure
Human Phenotype Ontology
Hypertrophic Cardiomyopathy
K-nearest Neighbors
Logistic regression
Logarithmic fold Change
Machine Learning
Minimum redundancy - Maximum Relevance
Next-generation Sequencing
Not a number
Peripheral Mononuclear Blood Cell
Root mean Squared Error
Random Forest
RNA-sequencing
Single Nucleotide Polymorphisms
SHapley Additive exPlanations
Transcripts per Million
Variant Effect Predictor
Whole Exome Sequencing
Whole Genome Sequencing
Xtreme Gradient Boosting
Tsao, C. W. et al. Heart Disease and Stroke Statistics-2022 update: a Report from the American Heart Association. Circulation . 145 (8), e153–e639 (2022).
PubMed Google Scholar
Krittanawong, C. et al. Artificial Intelligence and Cardiovascular Genetics. Life (Basel, Switzerland) 12 (2), 279 (2022).
ADS PubMed Google Scholar
Wung, S. F., Hickey, K. T., Taylor, J. Y. & Gallek, M. J. Cardiovascular genomics. J. Nurs. Scholarship: Official Publication Sigma Theta Tau Int. Honor Soc. Nurs. 45 (1), 60–68 (2013).
Google Scholar
Patel, K. K. et al. Genomic approaches to identify and investigate genes associated with atrial fibrillation and heart failure susceptibility. Hum. Genomics . 17 (1), 47 (2023).
PubMed PubMed Central Google Scholar
Ahmed, Z. Deciphering expression and variants in cardiovascular disease genes among heart failure population for precision medicine. ESC Heart Fail. 11 (1), 606–609 (2024).
Ahmed, Z. et al. Investigating genes associated with cardiovascular disease among heart failure patients for translational research and precision medicine. Clin. Translational Discovery , 3 (3), e206. (2023).
Zech, M. & Winkelmann, J. Next-generation sequencing and bioinformatics in rare movement disorders. Nat. Reviews Neurol. 20 (2), 114–126 (2024).
Byron, S. A., Van Keuren-Jensen, K. R., Engelthaler, D. M., Carpten, J. D. & Craig, D. W. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat. Rev. Genet. 17 (5), 257–271 (2016).
Abbas, M. & El-Manzalawy, Y. Machine learning based refined differential gene expression analysis of pediatric sepsis. BMC Med. Genom. 13 (1), 122 (2020).
Hamaguchi, Y., Zeng, C. & Hamada, M. Impact of human gene annotations on RNA-seq differential expression analysis. BMC Genom. 22 (1), 730 (2021).
Kaya, C. et al. Limitations of detecting genetic variants from the RNA sequencing data in tissue and fine-needle aspiration samples. Thyroid: Official J. Am. Thyroid Association . 31 (4), 589–595 (2021).
MathSciNet Google Scholar
Ellingford, J. M. et al. Recommendations for clinical interpretation of variants found in non-coding regions of the genome. Genome Med. 14 (1), 73 (2022).
Gonorazky, H. D. et al. Expanding the boundaries of RNA sequencing as a Diagnostic Tool for Rare mendelian disease. Am. J. Hum. Genet. 104 (3), 466–483 (2019).
Mhatre, I. et al. Functional mutation, splice, distribution, and divergence analysis of impactful genes associated with heart failure and other cardiovascular diseases. Sci. Rep. 13 (1), 16769 (2023).
ADS PubMed PubMed Central Google Scholar
Aung, N. et al. Genome-wide analysis of left ventricular image-derived phenotypes identifies fourteen loci Associated with Cardiac morphogenesis and heart failure development. Circulation . 140 (16), 1318–1330 (2019).
Bomba, L. et al. A. S., Whole-exome sequencing identifies rare genetic variants associated with human plasma metabolites. American journal of human genetics, 109(6), 1038–1054. (2022).
Xiao, W. et al. Toward best practice in cancer mutation detection with whole-genome and whole-exome sequencing. Nat. Biotechnol. 39 (9), 1141–1150 (2021).
Vadapalli, S., Abdelhalim, H., Zeeshan, S. & Ahmed, Z. Artificial intelligence and machine learning approaches using gene expression and variant data for personalized medicine. Brief. Bioinform. 23 (5), bbac191 (2022).
Weintraub, W. S. Role of Big Data in Cardiovascular Research. J. Am. Heart Association , 8 (14), e012791. (2019).
Armoundas, A. A. et al. Council on Lifelong Congenital Heart Disease and Heart Health in the Young; Council on Cardiovascular Radiology and Intervention; Council on Hypertension; Council on the Kidney in Cardiovascular Disease; and Stroke Council Use of Artificial Intelligence in Improving Outcomes in Heart Disease: A Scientific Statement From the American Heart Association. Circulation https://doi.org/10.1161/CIR.0000000000001201 (2024).
Article PubMed PubMed Central Google Scholar
Muse, E. D. & Topol, E. J. Transforming the cardiometabolic disease landscape: Multimodal AI-powered approaches in prevention and management. Cell metabolism, S1550-4131(24)00048 – 2. Advance online publication. (2024).
Nagarajan, V. D. et al. Artificial intelligence in the diagnosis and management of arrhythmias. Eur. Heart J. 42 (38), 3904–3916 (2021).
Kegerreis, B. et al. Machine learning approaches to predict lupus disease activity from gene expression data. Sci. Rep. 9 (1), 9617 (2019).
Maniruzzaman, M. et al. Statistical characterization and classification of colon microarray gene expression data using multiple machine learning paradigms. Comput. Methods Programs Biomed. 176 , 173–193 (2019).
Ahmed, Z., Zeeshan, S. & Liang, B. T. RNA-seq driven expression and enrichment analysis to investigate CVD genes with associated phenotypes among high-risk heart failure patients. Hum. Genomics . 15 (1), 67 (2021).
Venkat, V., Abdelhalim, H., DeGroat, W., Zeeshan, S. & Ahmed, Z. Investigating genes associated with heart failure, atrial fibrillation, and other cardiovascular diseases, and predicting disease using machine learning techniques for translational research and precision medicine. Genomics . 115 (2), 110584 (2023).
DeGroat, W., Venkat, V., Pierre-Louis, W., Abdelhalim, H. & Ahmed, Z. Hygieia: AI/ML pipeline integrating healthcare and genomics data to investigate genes associated with targeted disorders and predict disease. Softw. Impacts . 16 , 100493 (2023).
DeGroat, W. et al. Discovering biomarkers associated and predicting cardiovascular disease with high accuracy using a novel nexus of machine learning techniques for precision medicine. Sci. Rep. 14 (1), 1 (2024).
DeGroat, W. et al. IntelliGenes: a novel machine learning pipeline for biomarker discovery and predictive analysis using multi-genomic profiles. Bioinf. (Oxford England) . 39 (12), btad755 (2023).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550 (2014).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biol. 11 (10), R106 (2010).
Zhao, Y. et al. TPM, FPKM, or normalized counts? A comparative study of quantification measures for the analysis of RNA-seq data from the NCI patient-derived models repository. J. Translational Med. 19 (1), 269 (2021).
Radovic, M., Ghalwash, M., Filipovic, N. & Obradovic, Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinform. 18 (1), 9 (2017).
Ding, C. & Peng, H. Minimum redundancy feature selection from microarray gene expression data. J. Bioinform. Comput. Biol. 3 (2), 185–205 (2005).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A . 102 , 15545–15550 (2005).
The Gene. Ontology (GO) database and informatics resource. Nucleic Acids Res. 32 , D258–D261 (2004).
Gargano, M. A. et al. The human phenotype ontology in 2024: phenotypes around the world. Nucleic Acids Res. 52 , D1333–D1346 (2024).
Fishilevich, S. et al. D., GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database : the journal of biological databases and curation, 2017, bax028. (2017).
Schubach, M., Maass, T., Nazaretyan, L., Röner, S. & Kircher, M. CADD v1.7: using protein language models, regulatory CNNs and other nucleotide-level scores to improve genome-wide variant predictions. Nucleic Acids Res. 52 (D1), D1143–D1154 (2024).
Chen, S., Francioli, L. C., Goodrich, J. K., Collins, R. L., Kanai, M., Wang, Q., Alföldi, J., Watts, N. A., Vittal, C., Gauthier, L. D., Poterba, T., Wilson, M. W., Tarasova, Y., Phu, W., Grant, R., Yohannes, M. T., Koenig, Z., Farjoun, Y., Banks, E., Donnelly, S., … Karczewski, K. J. (2024). A genomic mutational constraint map using variation in 76,156 human genomes. Nature, 625(7993), 92–100.
Lundberg, S. M. & Lee, S. I. A unified approach to interpreting model predictions. Adv. Neural. Inf. Process. Syst. , 30. (2017).
Chonchol, M. & Nielson, C. Hemoglobin levels and coronary artery disease. Am. Heart J. 155 (3), 494–498 (2008).
Wei, J. et al. Key immune-related gene ITGB2 as a prognostic signature for acute myeloid leukemia. Annals Translational Med. 9 (17), 1386 (2021).
Zhu, L. et al. Identification of an association between coronary heart disease and ITGB2 methylation in peripheral blood by a case-control study. Clin. Chim. Acta . 552 , 117627 (2024).
Malinowski, D., Zawadzka, M., Safranow, K., Droździk, M. & Pawlik, A. SELL and GUCY1A1 gene polymorphisms in patients with unstable angina. Biomedicines . 10 (10), 2494 (2022).
Jin, J., Zhu, C., Wang, J., Zhao, X. & Yang, R. The association between ACTB methylation in peripheral blood and coronary heart disease in a case-control study. Front. Cardiovasc. Med. 9 , 972566 (2022).
Liu, C. et al. ACTB Methylation in blood as a potential marker for the pre-clinical detection of stroke: a prospective nested case-control study. Front. NeuroSci. 15 , 644943 (2021).
Mo, X. G. et al. NCF2, MYO1F, S1PR4, and FCN1 as potential noninvasive diagnostic biomarkers in patients with obstructive coronary artery: a weighted gene co-expression network analysis. J. Cell. Biochem. 120 (10), 18219–18235 (2019).
Chu, Y. et al. Identification of genes and key pathways underlying the pathophysiological association between nonalcoholic fatty liver disease and atrial fibrillation. BMC Med. Genom. 15 (1), 150 (2022).
Zicheng, H. et al. Association of circulating ICAM3 concentrations with severity and short-term outcomes of Acute ischemic stroke. Neurotox. Res. 39 (4), 1293–1299 (2021).
Abd Alla, J. et al. BBLN triggers CAMK2D pathology in mice under cardiac pressure overload and potentially in unrepaired hearts with tetralogy of Fallot. Nat. Cardiovasc. Res. 2 (11), 1044–1059 (2023).
Abd Alla, J. & Quitterer, U. Elevated BBLN levels as a cause of heart defects in tetralogy of Fallot. Nat. Cardiovasc. Res. 2 (11), 970–971 (2023).
Cao, J. & Yuan, L. Identification of key genes for hypertrophic cardiomyopathy using integrated network analysis of differential lncRNA and gene expression. Front. Cardiovasc. Med. 9 , 946229 (2022).
Tang, S., Liu, Y. & Liu, B. Integrated bioinformatics analysis reveals marker genes and immune infiltration for pulmonary arterial hypertension. Sci. Rep. 12 (1), 10154 (2022).
Ren, Z. et al. LC-MS/MS-Based absolute quantitation of hemoglobin subunits from dried blood spots reveals novel biomarkers for α-Thalassemia Silent Carriers. Anal. Chem. 95 (24), 9244–9251 (2023).
Joehanes, R. et al. Gene expression signatures of coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 33 (6), 1418–1426 (2013).
Prasad, K. Does HbA1cc play a role in the Development of Cardiovascular diseases? Curr. Pharm. Design . 24 (24), 2876–2882 (2018).
ADS Google Scholar
Ghaffari, S. et al. Association between HbA1c levels with severity of coronary artery disease and short-term outcomes of acute ST-elevation myocardial infarction in nondiabetic patients. Ther. Adv. Cardiovasc. Dis. 9 (5), 305–313 (2015).
Singer, D. E., Nathan, D. M., Anderson, K. M., Wilson, P. W. & Evans, J. C. Association of HbA1c with prevalent cardiovascular disease in the original cohort of the Framingham Heart Study. Diabetes . 41 (2), 202–208 (1992).
Ataga, K. I., Saraf, S. L. & Derebail, V. K. The nephropathy of sickle cell trait and sickle cell disease. Nat. Rev. Nephrol. 18 (6), 361–377 (2022).
Liu, W., Baker, S. S., Baker, R. D., Nowak, N. J. & Zhu, L. Upregulation of hemoglobin expression by oxidative stress in hepatocytes and its implication in nonalcoholic steatohepatitis. PloS One , 6 (9), e24363. (2011).
Aguilar, D. et al. Levels and change in Galectin-3 and Association with Cardiovascular events: the ARIC Study. J. Am. Heart Association , 9 (13), e015405. (2020).
Lubos, E., Loscalzo, J. & Handy, D. E. Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxid. Redox. Signal. 15 (7), 1957–1997 (2011).
Zhao, Y., Wang, H., Zhou, J. & Shao, Q. Glutathione peroxidase GPX1 and its dichotomous roles in Cancer. Cancers . 14 (10), 2560 (2022).
Kühn-Heid, E. C. D. et al. Selenium-Binding Protein 1 Indicates Myocardial Stress and Risk for Adverse Outcome in Cardiac Surgery. Nutrients, 11(9), 2005. (2019).
Chen, G. et al. Reduced selenium-binding protein 1 expression is associated with poor outcome in lung adenocarcinomas. J. Pathol. 202 (3), 321–329 (2004).
Zhang, Z. et al. Maternally inherited coronary heart disease is associated with a novel mitochondrial tRNA mutation. BMC Cardiovasc. Disord. 19 (1), 293 (2019).
Feng, W. & Han, S. lncRNA ADAMTS9-AS1/circFN1 competitively binds to miR-206 to elevate the expression of ACTB, thus inducing hypertrophic cardiomyopathy. Oxidative Med. Cell. Longev. 2022 , 1450610 (2022).
Lin, X., Zhou, Y. & Xue, L. Mitochondrial complex I subunit MT-ND1 mutations affect disease progression. Heliyon , 10 (7), e28808. (2024).
Straccia, G. et al. ACTB gene mutation in combined Dystonia-Deafness syndrome with parkinsonism: expanding the phenotype and highlighting the long-term GPi DBS outcome. Parkinsonism Relat. Disord. 104 , 3–6 (2022).
Zhang, Y. & He, Q. The role of SELENBP1 and its epigenetic regulation in carcinogenic progression. Front. Genet. 13 , 1027726 (2022).
Huang, N. et al. Role of NCF2 as a potential prognostic factor and immune infiltration indicator in hepatocellular carcinoma. Cancer Med. 12 (7), 8991–9004 (2023).
Tatewaki, M. et al. Constitutive overexpression of the L-selectin gene in fresh leukemic cells of adult T-cell leukemia that can be transactivated by human T-cell lymphotropic virus type 1 tax. Blood . 86 (8), 3109–3117 (1995).
Kim, K., Choi, E. Y., Ahn, H. M., Kim, D. G. & Kim, Y. J. Hemoglobin Subunit Theta 1 promotes proliferation by reducing reactive Oxygen species in Lung Adenocarcinoma. Cancers . 15 (23), 5504 (2023).
Alperin, J. B., Dow, P. A. & Petteway, M. B. Hemoglobin A2 levels in health and various hematologic disorders. Am. J. Clin. Pathol. 67 (3), 219–226 (1977).
Zhang, Q., Han, Q., Zi, J., Song, C. & Ge, Z. CD37 high expression as a potential biomarker and association with poor outcome in acute myeloid leukemia. Biosci. Rep. 40 (5), BSR20200008 (2020).
Mamegano, K. et al. Association of LILRA2 (ILT1, LIR7) splice site polymorphism with systemic lupus erythematosus and microscopic polyangiitis. Genes Immun. 9 (3), 214–223 (2008).
Che Yaacob, N. S. et al. Alpha-hemoglobin-stabilizing protein (AHSP): a modulatory factor in β-thalassemia. Int. J. Hematol. 111 , 352–359 (2020).
Jaing, T. H. et al. Molecular genetics of β-thalassemia: a narrative review. Medicine , 100 (45), e27522. (2021).
Verma, G., Rebholz-Schuhmann, D. & Madden, M. G. Enabling personalised disease diagnosis by combining a patient’s time-specific gene expression profile with a biomedical knowledge base. BMC Bioinform. 25 (1), 62 (2024).
Kim, S. I. et al. LYL1 gene amplification predicts poor survival of patients with uterine corpus endometrial carcinoma: analysis of the Cancer genome atlas data. BMC cancer . 18 (1), 494 (2018).
Kamino, Y. et al. HBD-2 is downregulated in oral carcinoma cells by DNA hypermethylation, and increased expression of hBD-2 by DNA demethylation and gene transfection inhibits cell proliferation and invasion. Oncol. Rep. 32 (2), 462–468 (2014).
Tian, T. et al. FKBP8 variants are risk factors for spina bifida. Hum. Mol. Genet. 29 (18), 3132–3144 (2020).
Tanaka, K. et al. Enhanced expression of mRNAs of antisecretory factor-1, gp96, DAD1 and CDC34 in human hepatocellular carcinomas. Biochim. Biophys. Acta . 1536 (1), 1–12 (2001).
Shi, X. et al. Combinatorial GxGxE CRISPR screen identifies SLC25A39 in mitochondrial glutathione transport linking iron homeostasis to OXPHOS. Nat. Commun. 13 (1), 2483 (2022).
Wang, X. et al. Association between structural brain features and gene expression by weighted gene co-expression network analysis in conversion from MCI to AD. Behav. Brain. Res. 410 , 113330 (2021).
Wan, J., Lv, J., Wang, C. & Zhang, L. RPS27 selectively regulates the expression and alternative splicing of inflammatory and immune response genes in thyroid cancer cells. Official Organ. Wroclaw Med. Univ. 31 (8), 889–901 (2022). Advances in clinical and experimental medicine .
Quan, W. et al. Identification of potential core genes in Parkinson’s Disease using Bioinformatics Analysis. Parkinson’s Disease . 2021 , 1690341 (2021).
Wright, C., Tawfik, M. A., Waisbourd, M. & Katz, L. J. Primary angle-closure glaucoma: an update. Acta Ophthalmol. 94 (3), 217–225 (2016).
Truebestein, L. & Leonard, T. A. Coiled-coils: the long and short of it. BioEssays : news and reviews in molecular. Cell. Dev. Biology . 38 (9), 903–916 (2016).
Ostermann, G., Weber, K. S., Zernecke, A., Schröder, A. & Weber, C. JAM-1 is a ligand of the beta(2) integrin LFA-1 involved in transendothelial migration of leukocytes. Nat. Immunol. 3 (2), 151–158 (2002).
Liu, C. et al. ACTB methylation in blood as a potential marker for the pre-clinical detection of stroke: a prospective nested case-control study. Front. NeuroSci. , 15 . (2021).
Chu, Y. et al. Identification of genes and key pathways underlying the pathophysiological association between nonalcoholic fatty liver disease and atrial fibrillation. BMC Med. Genomics . 15 , 150 (2022).
Zhang, X. et al. DYSF promotes monocyte activation in atherosclerotic cardiovascular disease as a DNA methylation-driven gene. Translational Res. 247 , 19–38 (2022).
van Leent, M. M. T. et al. Prosaposin mediates inflammation in atherosclerosis. Sci. Transl. Med. 13 (584), eabe1433 (2021).
Hwang, Y., Cornman, A. L., Kellogg, E. H., Ovchinnikov, S. & Girguis, P. R. Genomic language model predicts protein co-regulation and function. Nat. Commun. 15 (1), 2880 (2024).
Xie, Y. R. et al. Multiscale biochemical mapping of the brain through deep-learning-enhanced high-throughput mass spectrometry. Nat. Methods . 21 (3), 521–530 (2024).
Ahsan, M. U., Gouru, A., Chan, J., Zhou, W. & Wang, K. A signal processing and deep learning framework for methylation detection using Oxford Nanopore sequencing. Nat. Commun. 15 (1), 1448 (2024).
Osipov, A. et al. The Molecular Twin artificial-intelligence platform integrates multi-omic data to predict outcomes for pancreatic adenocarcinoma patients. Nat. cancer . 5 (2), 299–314 (2024).
Jiang, L. et al. Autosurv: interpretable deep learning framework for cancer survival analysis incorporating clinical and multi-omics data. Research square, rs.3.rs-2486756. (2023).
Aguilar, D., Bozkurt, B., Ramasubbu, K. & Deswal, A. Relationship of hemoglobin A1C and mortality in heart failure patients with diabetes. J. Am. Coll. Cardiol. 54 (5), 422–428 (2009).
Luo, P., Liu, X., Tang, Z. & Xiong, B. Decreased expression of HBA1 and HBB genes in acute myeloid leukemia patients and their inhibitory effects on growth of K562 cells. Hematol. (Amsterdam Netherlands) . 27 (1), 1003–1009 (2022).
Kühn, E. C. et al. Circulating levels of selenium-binding protein 1 (SELENBP1) are associated with risk for major adverse cardiac events and death. J. Trace Elem. Med. Biology: Organ. Soc. Minerals Trace Elem. (GMS) . 52 , 247–253 (2019).
Pol, A. et al. Mutations in SELENBP1, encoding a novel human methanethiol oxidase, cause extraoral halitosis. Nature genetics 50 (1), 120–129 (2018).
Suthahar, N. et al. Galectin-3 activation and inhibition in Heart failure and Cardiovascular Disease: an update. Theranostics . 8 (3), 593–609 (2018).
Song, S. et al. Overexpressed galectin-3 in pancreatic cancer induces cell proliferation and invasion by binding Ras and activating Ras signaling. PloS One , 7 (8), e42699. (2012).
Aureli, A., Cornò, D., Marziani, M., Gessani, B., Conti, L. & S., & Highlights on the role of Galectin-3 in Colorectal Cancer and the Preventive/Therapeutic potential of food-derived inhibitors. Cancers . 15 (1), 52 (2022).
Zifa, E. et al. A novel G3337A mitochondrial ND1 mutation related to cardiomyopathy co-segregates with tRNALeu(CUN) A12308G and tRNAThr C15946T mutations. Mitochondrion . 8 (3), 229–236 (2008).
Wang, Q. et al. Dysregulation of humoral immunity, iron homeostasis, and lipid metabolism is associated with multiple sclerosis progression. Multiple Scler. Relat. Disorders . 79 , 105020 (2023).
Singha, K., Sanchaisuriya, K., Fucharoen, G. & Fucharoen, S. Genetic and non-genetic factors affecting hemoglobin A2 expression in a large cohort of Thai individuals: implication for population screening for thalassemia. Am. J. Translational Res. 13 (10), 11632–11642 (2021).
Harthoorn-Lasthuizen, E. J., Lindemans, J. & Langenhuijsen, M. M. Influence of iron deficiency anaemia on haemoglobin A2 levels: possible consequences for beta-thalassaemia screening. Scand. J. Clin. Lab. Investig. 59 (1), 65–70 (1999).
El-Menshawy, N., Shahin, D. & Ghazi, H. F. Prognostic significance of the lymphoblastic leukemia-derived sequence 1 (LYL1) GeneExpression in Egyptian patients with AcuteMyeloid Leukemia. Turkish J. Haematol. : Official J. Turkish Soc. Haematol. 31 (2), 128–135 (2014).
Argentina, F., Suwarsa, O., Gunawan, H. & Berbudi, A. Gene expression of human Beta-Defensin-3 and Cathelicidin in the skin of Leprosy patients, Household contacts, and healthy individuals from Indonesia. Clin. Cosmet. Invest. Dermatology . 16 , 1485–1492 (2023).
Kanno, H. et al. Alpha hemoglobin stabilizing protein (AHSP) is a susceptibility gene to drug /infection-induced hemolytic anemia. Blood . 106 (11), 1678–1678 (2005).
Zhang, S. & Sun, Y. Targeting CDC34 E2 ubiquitin conjugating enzyme for lung cancer therapy. EBioMedicine . 54 , 102718 (2020).
Xu, H. et al. Gene expression profiling analysis of lung adenocarcinoma. Brazilian J. Med. Biol. Res. = Revista brasileira de pesquisas medicas e Biologicas , 49 (3), e4861. (2016).
Udhaya Kumar, S. et al. Analysis of differentially expressed genes and molecular pathways in familial hypercholesterolemia involved in atherosclerosis: a systematic and Bioinformatics Approach. Front. Genet. 11 , 734 (2020).
Rui, X., Shao, S., Wang, L. & Leng, J. Identification of recurrence marker associated with immune infiltration in prostate cancer with radical resection and build prognostic nomogram. BMC cancer . 19 (1), 1179 (2019).
Wang, R. et al. Loss of function mutations in RPL27 and RPS27 identified by whole-exome sequencing in Diamond-Blackfan anaemia. Br. J. Haematol. 168 (6), 854–864 (2015).
Gao, Y., Wang, Z., Yu, J. & Chen, L. Thyroid cancer and cardiovascular diseases: a mendelian randomization study. Front. Cardiovasc. Med. 11 , 1344515 (2024).
Namous, H. et al. ITGB2 is a central hub-gene associated with inflammation and early fibro-atheroma development in a swine model of atherosclerosis. Atherosclerosis plus . 54 , 30–41 (2023).
Tian, Y., Luo, Q., Huang, K., Sun, T. & Luo, S. Long noncoding RNA AC078850.1 induces NLRP3 inflammasome-mediated pyroptosis in atherosclerosis by upregulating ITGB2 transcription via transcription factor HIF-1α. Biomedicines . 11 (6), 1734 (2023).
Wu, J. et al. Identification of molecular signatures in acute myocardial infarction based on integrative analysis of proteomics and transcriptomics. Genomics . 115 (5), 110701 (2023).
Zu, L. et al. The Profile and clinical significance of ITGB2 expression in Non-small-cell Lung Cancer. J. Clin. Med. 11 (21), 6421 (2022).
Liu, H. et al. Correlation between ITGB2 expression and clinical characterization of glioma and the prognostic significance of its methylation in low-grade glioma (LGG). Front. Endocrinol. 13 , 1106120 (2023).
Li, C. et al. Identifying ITGB2 as a potential Prognostic Biomarker in Ovarian Cancer. Diagnostics (Basel Switzerland) . 13 (6), 1169 (2023).
Selim, A. M., Elsabagh, Y. A., El-Sawalhi, M. M., Ismail, N. A. & Senousy, M. A. Association of integrin-β2 polymorphism and expression with the risk of rheumatoid arthritis and osteoarthritis in Egyptian patients. BMC Med. Genom. 16 (1), 204 (2023).
Yang, S. et al. The ACTB Variants and Alcohol Drinking Confer Joint Effect to ischemic stroke in Chinese Han Population. J. Atheroscler. Thromb. 27 (3), 226–244 (2020).
Joseph, R., Srivastava, O. P. & Pfister, R. R. Downregulation of β-actin gene and human antigen R in human keratoconus. Investig. Ophthalmol. Vis. Sci. 53 (7), 4032–4041 (2012).
Li, H. et al. Associations of NADPH oxidase-related genes with blood pressure changes and incident hypertension: the GenSalt Study. J. Hum. Hypertens. 32 (4), 287–293 (2018).
Yang, S. et al. Potentially functional variants of ERAP1, PSMF1 and NCF2 in the MHC-I-related pathway predict non-small cell lung cancer survival. Cancer Immunol. Immunotherapy: CII . 70 (10), 2819–2833 (2021).
PubMed Central Google Scholar
Gu, P., Theiss, A., Han, J. & Feagins, L. A. Increased cell adhesion molecules, PECAM-1, ICAM-3, or VCAM-1, predict increased risk for flare in patients with quiescent inflammatory bowel disease. J. Clin. Gastroenterol. 51 (6), 522–527 (2017).
Xu-Monette, Z. Y. et al. Assessment of CD37 B-cell antigen and cell of origin significantly improves risk prediction in diffuse large B-cell lymphoma. Blood 128 (26), 3083–3100 (2016).
Zhang, J., Wang, L. & Jiang, M. Diagnostic value of sphingolipid metabolism-related genes CD37 and CXCL9 in nonalcoholic fatty liver disease. Medicine , 103 (8), e37185. (2024).
Dohm, A. et al. Identification of CD37, cystatin A, and IL-23A gene expression in association with brain metastasis: analysis of a prospective trial. Int. J. Biol. Mark. 34 (1), 90–97 (2019).
Fonseca, A. B. et al. The influence of innate and adaptative immune responses on the differential clinical outcomes of leprosy. Infect. Dis. Poverty . 6 (1), 5 (2017).
McMahon, M., Hahn, B. H. & Skaggs, B. J. Systemic lupus erythematosus and cardiovascular disease: prediction and potential for therapeutic intervention. Expert Rev. Clin. Immunol. 7 (2), 227–241 (2011).
Download references
Acknowledgements
We appreciate great support by Institute for Health, Health Care Policy and Aging Research (IFH), and Robert Wood Johnson Medical School (RWJMS), at Rutgers Health, and Pat and Jim Calhoun Cardiology Center, and Department of Genetics and Genome Sciences, at the UConn School of Medicine, UConn Health. We thank members and collaborators of Ahmed Lab at Rutgers (IFH, RWJMS, RBHS) for their support, participation, and contribution to this study. We appreciate all colleagues and institutions who provided direct and indirect insight and expertise that greatly assisted the research and development of this project. We acknowledge Rutgers Office of Advanced Research Computing (OARC) for providing access to the Amarel cluster and associated research computing resources.
Author information
William DeGroat, Habiba Abdelhalim, Elizabeth Peker, Neev Sheth and Rishabh Narayanan contributed equally.
Authors and Affiliations
Rutgers Institute for Health, Health Care Policy and Aging Research, Rutgers, The State University of New Jersey, 112 Paterson St, New Brunswick, NJ, 08901, USA
William DeGroat, Habiba Abdelhalim, Elizabeth Peker, Neev Sheth, Rishabh Narayanan & Zeeshan Ahmed
Department of Biomedical and Health Informatics, UMKC School of Medicine, 2411 Holmes Street, Kansas City, MO, 64108, USA
Saman Zeeshan
Pat and Jim Calhoun Cardiology Center, UConn Health, 263 Farmington Ave, Farmington, CT, USA
Bruce T. Liang
UConn School of Medicine, University of Connecticut, 263 Farmington Ave, Farmington, CT, USA
Bruce T. Liang & Zeeshan Ahmed
Department of Medicine, Division of Cardiovascular Disease and Hypertension, Robert Wood Johnson Medical School, Rutgers Health, 125 Paterson St, New Brunswick, NJ, 08901, USA
Zeeshan Ahmed
Rutgers Institute for Health, Health Care Policy and Aging Research, Rutgers University, 112 Paterson Street, New Brunswick, NJ, 08901, USA
You can also search for this author in PubMed Google Scholar
Contributions
Z.A. proposed, led, and supervised this study. Z.A. participated in conceptualization, project administration, funding acquisition, methodology, investigation, resource allocation, data duration, RNA-seq and WGS data processing, quality checking, downstream analysis. W.D. executed formal analysis, and R.N. tested and reproduced results. H.A., E.P., and N.S., participated in research, investigation, and validation of AI/ML results using state of the art literature. S.Z. guided post bioinformatics and AI/ML analysis and evaluated results. B.L. supported overall study including multi-omics data generation. All authors have participated in writing - original draft when Z.A performed review & editing. All authors have approved it for publication.
Corresponding author
Correspondence to Zeeshan Ahmed .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Ethical approval and consent to participate
Informed consent was obtained from all subjects. All human samples were used in accordance with relevant guidelines and regulations, and all experimental protocols were approved by the Institutional Review Board.
Supplementary Information
Accompanies this paper:
Supplementary material 1
Clinical sub-cohorts.
Supplementary material 2
CIGT format dataset.
Supplementary material 3: Metrics from machine learning classifiers.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary material 2, supplementary material 3, rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/ .
Reprints and permissions
About this article
Cite this article.
DeGroat, W., Abdelhalim, H., Peker, E. et al. Multimodal AI/ML for discovering novel biomarkers and predicting disease using multi-omics profiles of patients with cardiovascular diseases. Sci Rep 14 , 26503 (2024). https://doi.org/10.1038/s41598-024-78553-6
Download citation
Received : 07 June 2024
Accepted : 31 October 2024
Published : 03 November 2024
DOI : https://doi.org/10.1038/s41598-024-78553-6
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Machine learning
- Multi-omics
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.

COMMENTS
Creates a comprehensive academic literature review with scholarly resources based on a specific research topic. HyperWrite's AI Literature Review Generator is a revolutionary tool that automates the process of creating a comprehensive literature review. Powered by the most advanced AI models, this tool can search and analyze scholarly articles, books, and other resources to identify key themes ...
Using AI for literature review can significantly streamline your research process. Let's explore two methods: a multi-tool approach and using Elephas, an all-in-one assistant. Method 1: Using Multiple AI Tools for Literature Review. 1. Identify Your Research Topic and Keywords.
This paper presents a comprehensive review of the use of Artificial Intelligence (AI) in Systematic Literature Reviews (SLRs). A SLR is a rigorous and organised methodology that assesses and integrates prior research on a given topic. Numerous tools have been developed to assist and partially automate the SLR process. The increasing role of AI in this field shows great potential in providing ...
AI literature search and mapping - best AI tools for a literature review - elicit and more. Harnessing AI tools for literature reviews and mapping brings a new level of efficiency and precision to academic research. No longer do you have to spend hours looking in obscure research databases to find what you need! AI-powered tools like ...
In this essay, we focus on the use of AI-based tools in the conduct of literature reviews. Advancing knowledge in this area is particularly promising since (1) standalone review projects require substantial efforts over months and years (Larsen et al., 2019), (2) the volume of reviews published in IS journals has been rising steadily (Schryen et al., 2020), and (3) literature reviews involve ...
Using literature review AI tools can be a complete game changer in your research. They can make the literature review process smarter and hassle-free. Here are some practical benefits: Efficiency. AI tools for literature review can skim through tons of research papers and find the most relevant one for your topic in no time, thus saving you ...
Published on: December 22, 2023. Researching and writing outa literature review can be a daunting task, but AI-powered tools like Semantic Scholar, Research Rabbit, and Scite are revolutionising this process. These tools, leveraging artificial intelligence, automate the arduous task of sifting through mountains of academic papers, extracting ...
Schwa tz et al.,2020. What Is Semantic Scholar? Semantic Scholar is a free, AI-powered research tool for scientific literature, based at Ai2. Semantic Scholar uses groundbreaking AI and engineering to understand the semantics of scientific literature to help Scholars discover relevant research.
ATLAS.ti Web enables researchers to complete their literature review using AI tools and qualitative data analysis features. Targeted summaries from Paper Search 2.0 save time, allowing researchers to move forward more efficiently. The integrated tools make literature reviews more streamlined and less time-consuming. Future enhancements include ...
AI-Powered Literature Review: Faster, Deeper, Smarter. Enago Read simplifies literature reviews by delivering smart AI-driven summaries, key insights, real-time discovery, and a copilot that empowers you to master the literature with superhuman speed. Get Started. No credit card required. Used by researchers from more than 400 universities.
Welcome to Jenni AI, the ultimate tool for researchers and students. Our AI Literature Review Generator is designed to assist you in creating comprehensive, high-quality literature reviews, enhancing your academic and research endeavors. Say goodbye to writer's block and hello to seamless, efficient literature review creation.
Zotero. Zotero is an open-access, easy-to-use reference management tool designed to assist with the collection, organization, citation, and sharing of research sources. It serves as a personal research assistant for students, researchers, and academics, helping them manage their literature review process efficiently.
Over 2 million researchers have used Elicit. Researchers commonly use Elicit to: Speed up literature review; Find papers they couldn't find elsewhere; Automate systematic reviews and meta-analyses; Learn about a new domain; Elicit tends to work best for empirical domains that involve experiments and concrete results.
To make the process of finding specific information in big collections of documents quicker and easier — for example, in a literature review — search, we created the Lateral app, a new kind of AI-driven interface to help you organise, search through and save supporting quotes and information from collections of articles. Using techniques ...
Again, you can use AI Assist's power to support this step of your literature review. AI Assist analyzes the text and suggests subcodes while leaving the decision on whether you want to create the suggested sub-codes up to you. To create AI-generated subcode suggestions, open the context menu of a code and choose AI Assist > Suggest Subcodes. ...
A Step-by-Step Guide: How to Use ChatGPT for Writing a Literature Review. Step 1: Defining Your Research Objective Before diving into the literature review process, it is crucial to define your research objective. Clearly articulate the topic, research question, or hypothesis you aim to address through your literature review.
AI-POWERED RESEARCH ASSISTANT - finding papers, filtering study types, automating research flow, brainstorming, summarizing and more. "Elicit is a research assistant using language models like GPT-3 to automate parts of researchers' workflows. Currently, the main workflow in Elicit is Literature Review.
Recent advancements in artificial intelligence (AI) enable the rapid collection and organization of academic research with the push of a button, prompting a reevaluation of the role and value of human-generated systematic literature reviews (SLRs). In this editorial, we explore the value of the human element in producing SLRs, a method originally designed to reduce human bias in literature ...
Abstract. This manuscript presents a comprehensive review of the use of Artificial Intelligence (AI) in Systematic Literature Reviews (SLRs). A SLR is a rigorous and organised methodology that assesses and integrates prior research on a given topic. Numerous tools have been developed to assist and partially automate the SLR process.
Look no further! Our Literature Review Generator is a free AI writing tool designed to help you streamline your research and create a high-quality literature review. How it Works. Our Literature Review Generator uses advanced AI technology to analyze your input and generate a comprehensive literature review that is tailored to your specific needs.
AI-Powered Literature Review Generator. Generate high-quality literature reviews fast with our AI tool. Summarize papers, identify key themes, and synthesize conclusions with just a few clicks. The AI reviews thousands of sources to find the most relevant info for your topic.
A number of dental specialties, including pediatric dentistry, now use AI and its subsets, machine learning, and deep learning. The evolution of AI in healthcare has been linked to the creation of AI applications meant to support medical professionals in diagnosing patients and choosing the best course of treatment.
Table 3 Regulation of differentially expressed genes based on literature review. This table includes gene names, their regulation based on the log fold change (LFC), and their regulation based on ...